Identification and Strain Differentiation of Bacillus Species Using
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

The Food Poisoning Toxins of Bacillus Cereus
toxins Review The Food Poisoning Toxins of Bacillus cereus Richard Dietrich 1,†, Nadja Jessberger 1,*,†, Monika Ehling-Schulz 2 , Erwin Märtlbauer 1 and Per Einar Granum 3 1 Department of Veterinary Sciences, Faculty of Veterinary Medicine, Ludwig Maximilian University of Munich, Schönleutnerstr. 8, 85764 Oberschleißheim, Germany; [email protected] (R.D.); [email protected] (E.M.) 2 Department of Pathobiology, Functional Microbiology, Institute of Microbiology, University of Veterinary Medicine Vienna, 1210 Vienna, Austria; [email protected] 3 Department of Food Safety and Infection Biology, Faculty of Veterinary Medicine, Norwegian University of Life Sciences, P.O. Box 5003 NMBU, 1432 Ås, Norway; [email protected] * Correspondence: [email protected] † These authors have contributed equally to this work. Abstract: Bacillus cereus is a ubiquitous soil bacterium responsible for two types of food-associated gastrointestinal diseases. While the emetic type, a food intoxication, manifests in nausea and vomiting, food infections with enteropathogenic strains cause diarrhea and abdominal pain. Causative toxins are the cyclic dodecadepsipeptide cereulide, and the proteinaceous enterotoxins hemolysin BL (Hbl), nonhemolytic enterotoxin (Nhe) and cytotoxin K (CytK), respectively. This review covers the current knowledge on distribution and genetic organization of the toxin genes, as well as mechanisms of enterotoxin gene regulation and toxin secretion. In this context, the exceptionally high variability of toxin production between single strains is highlighted. In addition, the mode of action of the pore-forming enterotoxins and their effect on target cells is described in detail. The main focus of this review are the two tripartite enterotoxin complexes Hbl and Nhe, but the latest findings on cereulide and CytK are also presented, as well as methods for toxin detection, and the contribution of further putative virulence factors to the diarrheal disease. -

The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio In
microorganisms Review The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease Spase Stojanov 1,2, Aleš Berlec 1,2 and Borut Štrukelj 1,2,* 1 Faculty of Pharmacy, University of Ljubljana, SI-1000 Ljubljana, Slovenia; [email protected] (S.S.); [email protected] (A.B.) 2 Department of Biotechnology, Jožef Stefan Institute, SI-1000 Ljubljana, Slovenia * Correspondence: borut.strukelj@ffa.uni-lj.si Received: 16 September 2020; Accepted: 31 October 2020; Published: 1 November 2020 Abstract: The two most important bacterial phyla in the gastrointestinal tract, Firmicutes and Bacteroidetes, have gained much attention in recent years. The Firmicutes/Bacteroidetes (F/B) ratio is widely accepted to have an important influence in maintaining normal intestinal homeostasis. Increased or decreased F/B ratio is regarded as dysbiosis, whereby the former is usually observed with obesity, and the latter with inflammatory bowel disease (IBD). Probiotics as live microorganisms can confer health benefits to the host when administered in adequate amounts. There is considerable evidence of their nutritional and immunosuppressive properties including reports that elucidate the association of probiotics with the F/B ratio, obesity, and IBD. Orally administered probiotics can contribute to the restoration of dysbiotic microbiota and to the prevention of obesity or IBD. However, as the effects of different probiotics on the F/B ratio differ, selecting the appropriate species or mixture is crucial. The most commonly tested probiotics for modifying the F/B ratio and treating obesity and IBD are from the genus Lactobacillus. In this paper, we review the effects of probiotics on the F/B ratio that lead to weight loss or immunosuppression. -

Distribution and Characteristics of Bacillus Bacteria Associated with Hydrobionts and the Waters of the Peter the Great Bay, Sea of Japan I
ISSN 0026-2617, Microbiology, 2008, Vol. 77, No. 4, pp. 497–503. © Pleiades Publishing, Ltd., 2008. Original Russian Text © I.A. Beleneva, 2008, published in Mikrobiologiya, 2008, Vol. 77, No. 4, pp. 558–565. EXPERIMENTAL ARTICLES Distribution and Characteristics of Bacillus Bacteria Associated with Hydrobionts and the Waters of the Peter the Great Bay, Sea of Japan I. A. Beleneva1 Zhirmunskii Institute of Marine Biology, Far East Division, Russian Academy of Sciences, ul. Pal’chevskogo, 17, Vladivostok 690041, Russia Received May 28, 2007 Abstract—Bacilli of the species Bacillus subtilis, B. pumilus, B. mycoides, B. marinus and B. licheniformis (a total of 53 strains) were isolated from 15 invertebrate species and the water of the Vostok Bay, Peter the Great Bay, Sea of Japan. Bacilli were most often isolated from bivalves (22.7%) and sea cucumbers (18.9%); they occurred less frequently in sea urchins and starfish (13.2 and 7.5%, respectively). Most of bacilli strains were isolated from invertebrates inhabiting silted sediments. No Bacillus spp. strains were isolated from invertebrates inhabiting stony and sandy environments. The species diversity of bacilli isolated from marine objects under study was low. Almost all bacterial isolates were resistant to lincomycin. Unlike B. pumilus, B. subtilis isolates were mostly resistant to benzylpenicillin and ampicillin. Antibiotic sensitivity of B. licheniformis strains was variable (two strains were resistant to benzylpenicillin and oxacillin, while one was sensitive). A significant fraction of isolated bacilli contained pigments. Pigmented strains were more often isolated from seawater sam- ples, while colorless ones predominated within hydrobionts. B. subtilis colonies had the broadest range of co- lors. -

Chapter 11: Bacteria Bacterial Groups
Bacterial Groups u Most widely accepted taxonomic classification for bacteria is Bergey’s Manual of Systematic Bacteriology. u 5000 bacterial species identified, 3100 classified. Chapter 11: Bacteria u Bacteria are divided into four divisions (phyla) according to the characteristics of their cell walls. u Each division is divided into sections according to: u Gram stain reaction u Cell shape u Cell arrangements u Oxygen requirements u Motility u Nutritional and metabolic properties u Each section contains several genera. Four Divisions of Bacteria Classification of Bacteria Procaryotes Gram-Negative Division II Wall-Less Archaea Bacteria Bacteria Bacteria Bacteria (Gracilicutes) (Firmicutes) (Tenericutes) (Mendosicutes) Thin Cell Walls Thick cell Walls Lack cell walls Unusual cell walls Division I. Gram-Negative Bacteria Gram Negative Bacteria Spirochetes 1. Spirochetes u Helical shape. Flexible. u Contain two or more axial filaments (endoflagella). u Move in corkscrew pattern. u Medically important members: F Treponema pallidum: Syphilis F Borrelia spp.: Lyme disease, relapsing fever F Leptospira: Leptospirosis 1 Syphilis is Caused by a Spirochete Lyme Disease is Caused by a Spirochete Primary syphilitic chancre and secondary rash. Source: Tropical Medicine and Parasitology, 1997 Lyme Disease early lesion at tick bite site. Source: Medical Microbiology, 1998 2. Aerobic, Motile, Helical/Vibroid Gram- Negative Bacteria Gram Negative Bacteria u Rigid helical shape or curved rods. Aerobic, Motile, Helical/Vibroid u Lack axial filaments (endoflagella); have polar Gram-Negative Bacteria flagella instead. u Most are harmless aquatic organisms. u Genus Azospirillum fixes nitrogen in soil. u Genus Bdellovibrio attacks other bacteria. u Important pathogens include: F Campylobacter jejuni: Most common bacterial food- borne intestinal disease in the United States (2 million cases/year). -

Technical Method
J Clin Pathol: first published as 10.1136/jcp.38.9.1073 on 1 September 1985. Downloaded from J Clin Pathol 1985;38:1073-1084 Technical method Use of microwaves for acid and alcohol fast staining S HAFIZ, RC SPENCER, MARGARET LEE, organism depends on several factors but remains an HILARY GOOCH, BI DUERDEN Department important component in the identification of a ofMedical Microbiology, Sheffield University Medi- restricted group of organisms. cal School, Sheffield The technical problems associated with the classic Ziehl-Neelsen method include the dangers of heat- ing by flaming torch in laboratories in which volatile Certain bacteria that are characterised by a high organic solvents are used, the deposits that accumu- content of lipid in their cell walls cannot be stained late on the underside of the slide as a result of heat- by simple stains, and either heat or prolonged con- ing, and the crystalline deposits of stain that collect tact is required to drive the stain into the cells; once on the film as a result of evaporation and drying. stained they resist decolourisation by mineral acids This method also requires 20-30 minutes of or acid and alcohol. These organisms are designated laboratory time. Numerous minor modifications acid fast or acid and alcohol fast. They include have been made to the Ziehl-Neelsen method by Mycobacterium tuberculosis and related mycobac- increasing the concentration of carbolic magenta or teria, Mycobacterium leprae, and certain of the by cold staining techniques,'0-'2 but the basic prob- actinomycetes. Koch first stained the tubercle bacil- lems remain. lus by immersion in an alkaline solution of Some of the problems may be overcome by the copyright. -

Araştırma Isolation and Characterization of Bacillus
Akademik Ziraat Dergisi 7(1):21-28 (2018) Araştırma ISSN: 2147-6403 DOI: http://dx.doi.org/10.29278/azd.440586 (Research) Isolation and characterization of Bacillus megaterium isolates from dead pentatomids and their insecticidal activity to Palomena prasina nymphs Hasan Murat AKSOY1, Celal TUNCER1, Islam SARUHAN1, Ismail ERPER1, Murat OZTURK1, Izzet AKCA1 1Ondokuz Mayis University, Faculty of Agriculture, Department of Plant Protection, SAMSUN , Kabul tarihi 06 Nisan 2018 Sorumlu yazar: , e-posta:[email protected] Alınış tarihi: 19 Aralık 2017 İslam SARUHAN Abstract Ölü pentatomidlerden Bacillus megaterium Bacillus megaterium isolates, were demonstrated to izolatlarının izolasyonu, karekterizasyonu ve be efficient biocontrol agents against the green Palomena prasina nimflerine insektisital etkisi shield bug (Palomena prasina Pentatomidae). Firstly hazelnut orchards were Öz surveyed and four B. megateriumL., isolatesHeteroptera: were Bacillus megaterium obtained from P. prasina. The morphological, Palomena prasina physiological and biochemical characteristics of B. Bu çalışma, izolatlarının fındık megaterium isolates were determined according to kokarcasına ( L., Heteroptera: the standardized methodology. Additionally 16S Pentatomidae) karşıP. etkinprasina biyokontrol Bacillusajanları to megateriumolduğu gösterilmiştir. Öncelikle fındıkB. bahçelerindemegaterium survey yapılarak, 'dan dört rRNA gene sequence analyse was performed gene confirmed that isolates Sa-1, Sa-5, SAk-2 and izolatı elde edilmiştir. determine the isolates. Analysis of the 16S rRNA SAkc-19 are B. megaterium izolatlarının morfolojik, fizyolojik ve biyokimyasal homology to the type strains of B. megaterium. özellikleri standart tanı yöntemlerine göre Effectiveness of these isolates, withwas tested100% againstsequence P. sonucu;belirlenmiştir. Sa-1, Sa Ayrıca-5, SAk izolatların-2 ve SAkc tanısı- için 16S rRNAB. prasina nymphs in laboratory, at 25±1°C and 70±5 megateriumgen dizi analizi yapılmıştır. -

Common Commensals
Common Commensals Actinobacterium meyeri Aerococcus urinaeequi Arthrobacter nicotinovorans Actinomyces Aerococcus urinaehominis Arthrobacter nitroguajacolicus Actinomyces bernardiae Aerococcus viridans Arthrobacter oryzae Actinomyces bovis Alpha‐hemolytic Streptococcus, not S pneumoniae Arthrobacter oxydans Actinomyces cardiffensis Arachnia propionica Arthrobacter pascens Actinomyces dentalis Arcanobacterium Arthrobacter polychromogenes Actinomyces dentocariosus Arcanobacterium bernardiae Arthrobacter protophormiae Actinomyces DO8 Arcanobacterium haemolyticum Arthrobacter psychrolactophilus Actinomyces europaeus Arcanobacterium pluranimalium Arthrobacter psychrophenolicus Actinomyces funkei Arcanobacterium pyogenes Arthrobacter ramosus Actinomyces georgiae Arthrobacter Arthrobacter rhombi Actinomyces gerencseriae Arthrobacter agilis Arthrobacter roseus Actinomyces gerenseriae Arthrobacter albus Arthrobacter russicus Actinomyces graevenitzii Arthrobacter arilaitensis Arthrobacter scleromae Actinomyces hongkongensis Arthrobacter astrocyaneus Arthrobacter sulfonivorans Actinomyces israelii Arthrobacter atrocyaneus Arthrobacter sulfureus Actinomyces israelii serotype II Arthrobacter aurescens Arthrobacter uratoxydans Actinomyces meyeri Arthrobacter bergerei Arthrobacter ureafaciens Actinomyces naeslundii Arthrobacter chlorophenolicus Arthrobacter variabilis Actinomyces nasicola Arthrobacter citreus Arthrobacter viscosus Actinomyces neuii Arthrobacter creatinolyticus Arthrobacter woluwensis Actinomyces odontolyticus Arthrobacter crystallopoietes -

Use of the Diagnostic Bacteriology Laboratory: a Practical Review for the Clinician
148 Postgrad Med J 2001;77:148–156 REVIEWS Postgrad Med J: first published as 10.1136/pmj.77.905.148 on 1 March 2001. Downloaded from Use of the diagnostic bacteriology laboratory: a practical review for the clinician W J Steinbach, A K Shetty Lucile Salter Packard Children’s Hospital at EVective utilisation and understanding of the Stanford, Stanford Box 1: Gram stain technique University School of clinical bacteriology laboratory can greatly aid Medicine, 725 Welch in the diagnosis of infectious diseases. Al- (1) Air dry specimen and fix with Road, Palo Alto, though described more than a century ago, the methanol or heat. California, USA 94304, Gram stain remains the most frequently used (2) Add crystal violet stain. USA rapid diagnostic test, and in conjunction with W J Steinbach various biochemical tests is the cornerstone of (3) Rinse with water to wash unbound A K Shetty the clinical laboratory. First described by Dan- dye, add mordant (for example, iodine: 12 potassium iodide). Correspondence to: ish pathologist Christian Gram in 1884 and Dr Steinbach later slightly modified, the Gram stain easily (4) After waiting 30–60 seconds, rinse with [email protected] divides bacteria into two groups, Gram positive water. Submitted 27 March 2000 and Gram negative, on the basis of their cell (5) Add decolorising solvent (ethanol or Accepted 5 June 2000 wall and cell membrane permeability to acetone) to remove unbound dye. Growth on artificial medium Obligate intracellular (6) Counterstain with safranin. Chlamydia Legionella Gram positive bacteria stain blue Coxiella Ehrlichia Rickettsia (retained crystal violet). -

Bacillus Subtilis: Model Organism for Cellular Development, and Industrial Workhorse
MICROBE PROFILE Errington and Aart, Microbiology 2020;166:425–427 DOI 10.1099/mic.0.000922 Microbe Profile: Bacillus subtilis: model organism for cellular development, and industrial workhorse Jeffery Errington* and Lizah T van der Aart Graphical abstract Life cycle, environmental importance and industrial applications of B. subtilis. DNA and life cycle: the laboratory strain of B. subtilis is naturally transformable and, in the typical example illustrated, a foreign DNA segment ‘insert’ is integrated into the amyE genetic locus by double crossover homologus recombination. A crucial facet of the life cycle of most B. subtilis and most Firmicutes is their ability to switch from a classical binary fission, with equal segregation of sister chromosomes,to endospore formation. The resultant asymmetrical division generates small prespore (red) and larger mother- cell (green) compartments with different patterns of transcription. The tough endospore that results can remain dormant for a long period of time before germinating to resume vegetative growth. Environmental interactions: B. subtilis is typically found in association with plants as both an epiphyte and also within the rhizosphere. In some parts of the world batches of spores are used extensively for plant protection in the form of a seed dressing. B. subtilis has also been studied extensively as a model system for biofilm formation, switching classically between planktonic and sessile states. Industrial applications: B. subtilis and closely related organisms are responsible for huge levels of production of hydrolytic commodity enzymes, particularly proteases and amylases. They are also popular in probiotic formulations and can be engineered for production of fine chemicals, such as the vitamin, riboflavin. -

Cell Structure and Function in the Bacteria and Archaea
4 Chapter Preview and Key Concepts 4.1 1.1 DiversityThe Beginnings among theof Microbiology Bacteria and Archaea 1.1. •The BacteriaThe are discovery classified of microorganismsinto several Cell Structure wasmajor dependent phyla. on observations made with 2. theThe microscope Archaea are currently classified into two 2. •major phyla.The emergence of experimental 4.2 Cellscience Shapes provided and Arrangements a means to test long held and Function beliefs and resolve controversies 3. Many bacterial cells have a rod, spherical, or 3. MicroInquiryspiral shape and1: Experimentation are organized into and a specific Scientificellular c arrangement. Inquiry in the Bacteria 4.31.2 AnMicroorganisms Overview to Bacterialand Disease and Transmission Archaeal 4.Cell • StructureEarly epidemiology studies suggested how diseases could be spread and 4. Bacterial and archaeal cells are organized at be controlled the cellular and molecular levels. 5. • Resistance to a disease can come and Archaea 4.4 External Cell Structures from exposure to and recovery from a mild 5.form Pili allowof (or cells a very to attach similar) to surfacesdisease or other cells. 1.3 The Classical Golden Age of Microbiology 6. Flagella provide motility. Our planet has always been in the “Age of Bacteria,” ever since the first 6. (1854-1914) 7. A glycocalyx protects against desiccation, fossils—bacteria of course—were entombed in rocks more than 3 billion 7. • The germ theory was based on the attaches cells to surfaces, and helps observations that different microorganisms years ago. On any possible, reasonable criterion, bacteria are—and always pathogens evade the immune system. have been—the dominant forms of life on Earth. -

Engineering Bacillus Megaterium for Production of Functional Intracellular Materials Katrin Grage1, Paul Mcdermott2 and Bernd H
Grage et al. Microb Cell Fact (2017) 16:211 DOI 10.1186/s12934-017-0823-5 Microbial Cell Factories RESEARCH Open Access Engineering Bacillus megaterium for production of functional intracellular materials Katrin Grage1, Paul McDermott2 and Bernd H. A. Rehm3* Abstract Background: Over the last 10–15 years, a technology has been developed to engineer bacterial poly(3-hydroxybu- tyrate) (PHB) inclusions as functionalized beads, for applications such as vaccines, diagnostics and enzyme immobili- zation. This has been achieved by translational fusion of foreign proteins to the PHB synthase (PhaC). The respective fusion protein mediates self-assembly of PHB inclusions displaying the desired protein function. So far, beads have mainly been produced in recombinant Escherichia coli, which is problematic for some applications as the lipopolysac- charides (LPS) co-purifed with such inclusions are toxic to humans and animals. Results: In this study, we have bioengineered the formation of functional PHB inclusions in the Gram-positive bac- terium Bacillus megaterium, an LPS-free and established industrial production host. As B. megaterium is a natural PHB producer, the PHB-negative strain PHA05 was used to avoid any background PHB production. Plasmid-mediated T7 promoter-driven expression of the genes encoding β-ketothiolase (phaA), acetoacetyl-CoA-reductase (phaB) and PHB synthase (phaC) enabled PHB production in B. megaterium PHA05. To produce functionalized PHB inclusions, the N- and C-terminus of PhaC was fused to four and two IgG binding Z-domains from Staphylococcus aureus, respectively. The ZZ-domain PhaC fusion protein was strongly overproduced at the surface of the PHB inclusions and the cor- responding isolated ZZ-domain displaying PHB beads were found to purify IgG with a binding capacity of 40–50 mg IgG/g beads. -
Bacillus Cereus
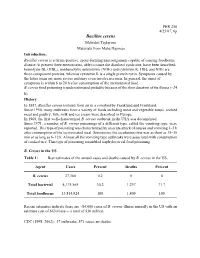
PHR 250 4/25/07, 6p Bacillus cereus Mehrdad Tajkarimi Materials from Maha Hajmeer Introduction: Bacillus cereus is a Gram-positive, spore-forming microorganism capable of causing foodborne disease At present three enterotoxins, able to cause the diarrheal syndrome, have been described: hemolysin BL (HBL), nonhemolytic enterotoxin (NHE) and cytotoxin K. HBL and NHE are three-component proteins, whereas cytotoxin K is a single protein toxin. Symptoms caused by the latter toxin are more severe and may even involve necrosis. In general, the onset of symptoms is within 6 to 24 h after consumption of the incriminated food. B. cereus food poisoning is underestimated probably because of the short duration of the illness (~24 h). History In 1887, Bacillus cereus isolated from air in a cowshed by Frankland and Frankland. Since 1950, many outbreaks from a variety of foods including meat and vegetable soups, cooked meat and poultry, fish, milk and ice cream were described in Europe. In 1969, the first well-characterized B. cereus outbreak in the USA was documented. Since 1971, a number of B. cereus poisonings of a different type, called the vomiting type, were reported. This type of poisoning was characterized by an acute attack of nausea and vomiting 1–5 h after consumption of the incriminated meal. Sometimes, the incubation time was as short as 15–30 min or as long as 6–12 h. Almost all the vomiting type outbreaks were associated with consumption of cooked rice. This type of poisoning resembled staphylococcal food poisoning. B. Cereus in the US Table 1: Best estimates of the annual cases and deaths caused by B.