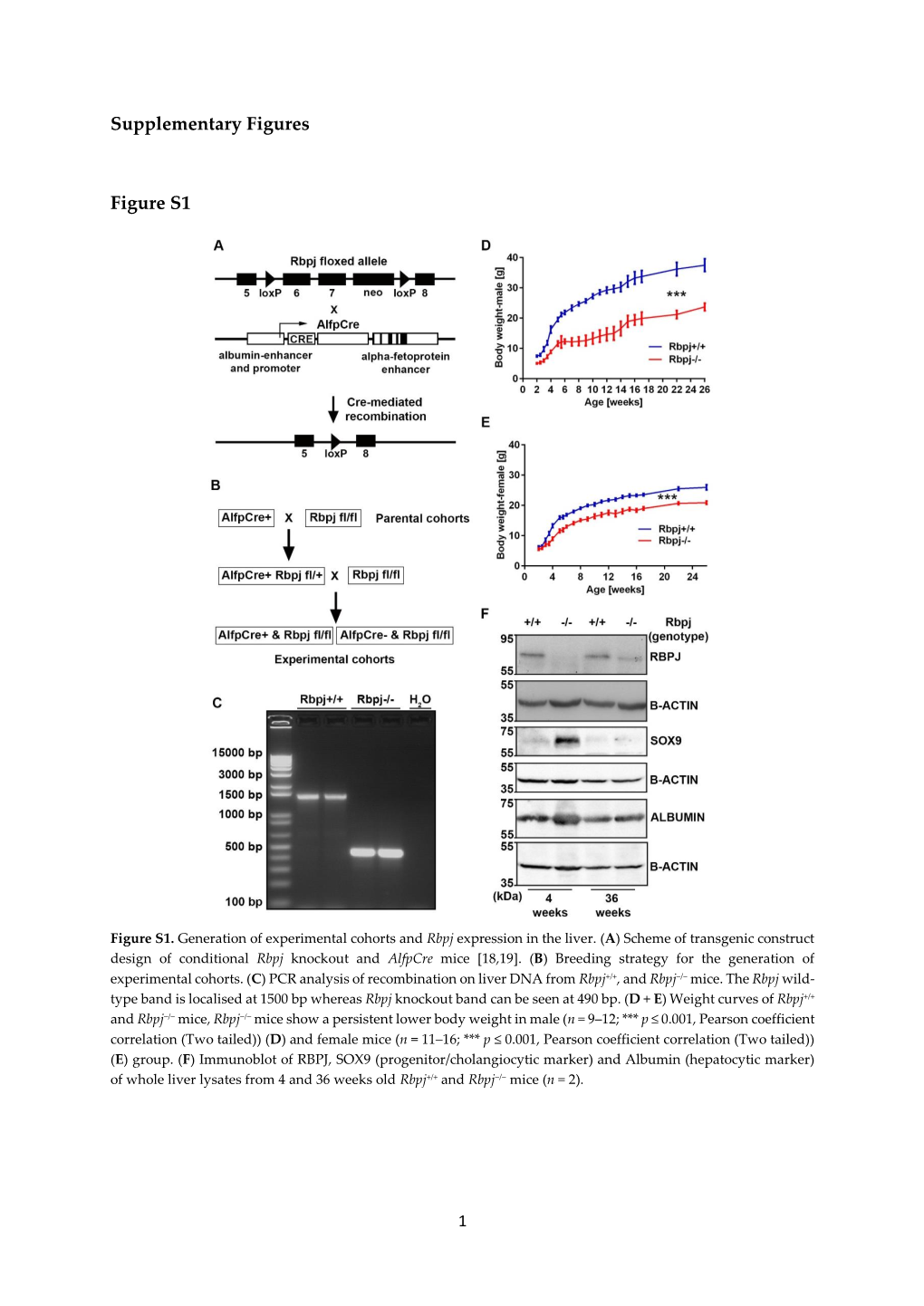
Supplementary Figures Figure S1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Methylation of the Homeobox Gene, HOPX, Is Frequently Detected in Poorly Differentiated Colorectal Cancer
ANTICANCER RESEARCH 31: 2889-2892 (2011) Methylation of the Homeobox Gene, HOPX, Is Frequently Detected in Poorly Differentiated Colorectal Cancer YOSHIKUNI HARADA, KAZUHIRO KIJIMA, KAZUKI SHINMURA, MAKIKO SAKATA, KAZUMA SAKURABA, KAZUAKI YOKOMIZO, YOUHEI KITAMURA, ATSUSHI SHIRAHATA, TETSUHIRO GOTO, HIROKI MIZUKAMI, MITSUO SAITO, GAKU KIGAWA, HIROSHI NEMOTO and KENJI HIBI Gastroenterological Surgery, Showa University Fujigaoka Hospital, 1-30 Fujigaoka, Aoba-ku, Yokohama 227-8501, Japan Abstract. Background: Homeodomein only protein x pathogenesis of colorectal cancer (4-8). It is also known that (HOPX) gene methylation has frequently been detected in gene promoter hypermethylation is involved in the cancer tissues. The methylation status of the HOPX gene in development and progression of cancer (9). An investigation colorectal cancer was examined and compared to the of genetic changes is important in order to clarify the clinocopathological findings. Materials and Methods: tumorigenic pathway of colorectal cancer (10). Eighty-nine tumor samples and corresponding normal tissues The homeodomain only protein x (HOPX) gene, also were obtained from colorectal cancer patients who known as NECC1 (not expressed in choriocarcinoma clone underwent surgery at our hospital. The methylation status of 1), LAGY (lung cancer-associated gene Y), and OB1 (odd the HOPX gene in these samples was examined by homeobox 1 protein), was initially identified as an essential quantitative methylation-specific PCR (qMSP). Subsequently, gene for the modulation of cardiac growth and development the clinicopathological findings were correlated with the (11). Three spliced transcript variants, HOPX-α, HOPX-β methylation status of the HOPX gene. Results: HOPX gene and HOPX-γ, encode the same protein, which contains a methylation was found in 46 (52%) out of the 89 colorectal putative homeodomain motif that acts as an adapter protein carcinomas, suggesting that it was frequently observed in to mediate transcriptional repression (12). -

Deciphering the Functions of Ets2, Pten and P53 in Stromal Fibroblasts in Multiple
Deciphering the Functions of Ets2, Pten and p53 in Stromal Fibroblasts in Multiple Breast Cancer Models DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Julie Wallace Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2013 Dissertation Committee: Michael C. Ostrowski, PhD, Advisor Gustavo Leone, PhD Denis Guttridge, PhD Dawn Chandler, PhD Copyright by Julie Wallace 2013 Abstract Breast cancer is the second most common cancer in American women, and is also the second leading cause of cancer death in women. It is estimated that nearly a quarter of a million new cases of invasive breast cancer will be diagnosed in women in the United States this year, and approximately 40,000 of these women will die from breast cancer. Although death rates have been on the decline for the past decade, there is still much we need to learn about this disease to improve prevention, detection and treatment strategies. The majority of early studies have focused on the malignant tumor cells themselves, and much has been learned concerning mutations, amplifications and other genetic and epigenetic alterations of these cells. However more recent work has acknowledged the strong influence of tumor stroma on the initiation, progression and recurrence of cancer. Under normal conditions this stroma has been shown to have protective effects against tumorigenesis, however the transformation of tumor cells manipulates this surrounding environment to actually promote malignancy. Fibroblasts in particular make up a significant portion of this stroma, and have been shown to impact various aspects of tumor cell biology. -

(12) United States Patent (10) Patent No.: US 9,109,232 B2 Schwartz Et Al
US009 109232B2 (12) United States Patent (10) Patent No.: US 9,109,232 B2 Schwartz et al. (45) Date of Patent: Aug. 18, 2015 (54) ETS2 AND MESP1 GENERATE CARDIAC Islas et al., Transcription factors ETS2 and MESP1 transdifferentiate PROGENITORS FROM FIBROBLASTS human dermal fibroblasts into cardiac progenitors; PNAS, Published online before print Jul. 23, 2012, doi: 10.1073/pnas. 11202991.09, 2012.* (71) Applicants: University of Houston, Houston, TX Melotti et al., Ets-2 and c-Myb Act Independently in Regulating (US); Texas Heart Institute, Houston, Expression of the Hematopoietic StemCell Antigen CD34:JBC, vol. TX (US); The Texas A&M University 269, No. 41, pp. 25303-25309, 1994.* System, College Station, TX (US) Takahashi et al., Induction of pluripotent stem cells from adulthuman fibroblasts by defined factors; Cell, vol. 131, pp. 861-872, 2007.* Naldini et al. In vivo gene delivery and stable transduction of (72) Inventors: Robert J. Schwartz, Houston, TX (US); nondividing cells by a lentiviral vector; Science, vol. 272, pp. 263 Vladimir N. Potaman, Houston, TX 267, 1996.* (US); Jose Francisco Islas, Houston, TX European Patent Office; Office Action; European Application No. (US) 117114082; Jul 22, 2013. European Patent Office; Response to Office Action; European Appli (73) Assignees: University of Houston, Houston, TX cation No. 11711408.2; Aug. 30, 2013. Chinese Patent Office; Office Action; Chinese Patent Application No. (US); Texas Heart Institute, Houston, 2011800 19561.0; Sep. 18, 2013. TX (US); The Texas A&M University Chinese Patent Office; Office Action (English translation); Chinese System, College Station, TX (US) Patent Application No. 2011800 19561.0; Sep. -

Overexpresssion of Tyrosine Kinase Discoidin Domain Receptor I (DDR1 ) in Transitional Cell Carcinoma
Overexpresssion of Tyrosine Kinase Discoidin domain receptor I (DDR1 ) in Transitional cell Carcinoma Szu-Ting Chen,* and Shie-Liang Hsieh* Institute and Department of Microbiology and Immunology, National Yang-Ming University, Taipei, Taiwan; To whom proofs are to be sent: Shie-Liang Hsieh, Department and Institute of Microbiology and Immunology, National Yang-Ming University, Shih-Pai, Taipei 112, Taiwan. E-mail address: [email protected] Telephone number: 886-2-28267161 Fax number: 886-2-28277933 INTRODUCTION DDR1, discoidin domain receptor 1, belongs to the novel subfamily of tyrosine kinase receptor, which forms homodimer upon ligand engagement. DDR1 is distinguished from other receptor tyrosine kinase by the discoidin domain in their extracellular domain, which is a homology region originally identified in Dictyostelium discoideum (slime mold) protein, discoidin I, and involves in cells aggregation. Discoidin-1 has binding specificity toward galactose and N-acetyl galactosamine and is essential for slime mold cells adhesion, migration and aggregation during its development, suggesting that DDR1 shared similar biologically function in mammalian [1-3]. DDR1 has been found mainly distributed and in human tissue epithelia, such as kidney, breast, lung [3], bronchial [4] and keratinocytes [5]. Furthermore, DDR1 has been also reported its expression in immune system like the monocyte-derived dendritic cells, annotated as CD167 [6] and macrophage[7]. However, recently, the overexpression of DDR1 has been detected in several human cancers, such as primary breast cancer [1, 8, 9] ovarian[10, 11], brain[12], esophageal cancer [13], and TCC (our data unpublished) in which raising the possibility that DDR1 may play a important role in tumorigenesis [14]. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Activated Peripheral-Blood-Derived Mononuclear Cells
Transcription factor expression in lipopolysaccharide- activated peripheral-blood-derived mononuclear cells Jared C. Roach*†, Kelly D. Smith*‡, Katie L. Strobe*, Stephanie M. Nissen*, Christian D. Haudenschild§, Daixing Zhou§, Thomas J. Vasicek¶, G. A. Heldʈ, Gustavo A. Stolovitzkyʈ, Leroy E. Hood*†, and Alan Aderem* *Institute for Systems Biology, 1441 North 34th Street, Seattle, WA 98103; ‡Department of Pathology, University of Washington, Seattle, WA 98195; §Illumina, 25861 Industrial Boulevard, Hayward, CA 94545; ¶Medtronic, 710 Medtronic Parkway, Minneapolis, MN 55432; and ʈIBM Computational Biology Center, P.O. Box 218, Yorktown Heights, NY 10598 Contributed by Leroy E. Hood, August 21, 2007 (sent for review January 7, 2007) Transcription factors play a key role in integrating and modulating system. In this model system, we activated peripheral-blood-derived biological information. In this study, we comprehensively measured mononuclear cells, which can be loosely termed ‘‘macrophages,’’ the changing abundances of mRNAs over a time course of activation with lipopolysaccharide (LPS). We focused on the precise mea- of human peripheral-blood-derived mononuclear cells (‘‘macro- surement of mRNA concentrations. There is currently no high- phages’’) with lipopolysaccharide. Global and dynamic analysis of throughput technology that can precisely and sensitively measure all transcription factors in response to a physiological stimulus has yet to mRNAs in a system, although such technologies are likely to be be achieved in a human system, and our efforts significantly available in the near future. To demonstrate the potential utility of advanced this goal. We used multiple global high-throughput tech- such technologies, and to motivate their development and encour- nologies for measuring mRNA levels, including massively parallel age their use, we produced data from a combination of two distinct signature sequencing and GeneChip microarrays. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Prospective Isolation of NKX2-1–Expressing Human Lung Progenitors Derived from Pluripotent Stem Cells
The Journal of Clinical Investigation RESEARCH ARTICLE Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells Finn Hawkins,1,2 Philipp Kramer,3 Anjali Jacob,1,2 Ian Driver,4 Dylan C. Thomas,1 Katherine B. McCauley,1,2 Nicholas Skvir,1 Ana M. Crane,3 Anita A. Kurmann,1,5 Anthony N. Hollenberg,5 Sinead Nguyen,1 Brandon G. Wong,6 Ahmad S. Khalil,6,7 Sarah X.L. Huang,3,8 Susan Guttentag,9 Jason R. Rock,4 John M. Shannon,10 Brian R. Davis,3 and Darrell N. Kotton1,2 2 1Center for Regenerative Medicine, and The Pulmonary Center and Department of Medicine, Boston University School of Medicine, Boston, Massachusetts, USA. 3Center for Stem Cell and Regenerative Medicine, Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center, Houston, Texas, USA. 4Department of Anatomy, UCSF, San Francisco, California, USA. 5Division of Endocrinology, Diabetes and Metabolism, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, Massachusetts, USA. 6Department of Biomedical Engineering and Biological Design Center, Boston University, Boston, Massachusetts, USA. 7Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts, USA. 8Columbia Center for Translational Immunology & Columbia Center for Human Development, Columbia University Medical Center, New York, New York, USA. 9Department of Pediatrics, Monroe Carell Jr. Children’s Hospital, Vanderbilt University, Nashville, Tennessee, USA. 10Division of Pulmonary Biology, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA. It has been postulated that during human fetal development, all cells of the lung epithelium derive from embryonic, endodermal, NK2 homeobox 1–expressing (NKX2-1+) precursor cells. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

UNIVERSITY of CALIFORNIA, IRVINE Combinatorial Regulation By
UNIVERSITY OF CALIFORNIA, IRVINE Combinatorial regulation by maternal transcription factors during activation of the endoderm gene regulatory network DISSERTATION submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biological Sciences by Kitt D. Paraiso Dissertation Committee: Professor Ken W.Y. Cho, Chair Associate Professor Olivier Cinquin Professor Thomas Schilling 2018 Chapter 4 © 2017 Elsevier Ltd. © 2018 Kitt D. Paraiso DEDICATION To the incredibly intelligent and talented people, who in one way or another, helped complete this thesis. ii TABLE OF CONTENTS Page LIST OF FIGURES vii LIST OF TABLES ix LIST OF ABBREVIATIONS X ACKNOWLEDGEMENTS xi CURRICULUM VITAE xii ABSTRACT OF THE DISSERTATION xiv CHAPTER 1: Maternal transcription factors during early endoderm formation in 1 Xenopus Transcription factors co-regulate in a cell type-specific manner 2 Otx1 is expressed in a variety of cell lineages 4 Maternal otx1 in the endodermal conteXt 5 Establishment of enhancers by maternal transcription factors 9 Uncovering the endodermal gene regulatory network 12 Zygotic genome activation and temporal control of gene eXpression 14 The role of maternal transcription factors in early development 18 References 19 CHAPTER 2: Assembly of maternal transcription factors initiates the emergence 26 of tissue-specific zygotic cis-regulatory regions Introduction 28 Identification of maternal vegetally-localized transcription factors 31 Vegt and OtX1 combinatorially regulate the endodermal 33 transcriptome iii