Precipitation of RNA with Ethanol
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Methanogenesis Rates in Acetate and Nitrate Amended Anoxic Slurries
Methanogenesis rates in acetate and nitrate amended anoxic slurries BIOS 35502: Practicum in Environmental Field Biology Patrick Revord Advisor: William West 2011 Abstract With increasing urbanization and land use changes, pollution of lakes and wetland ecosystems is imminent. Any influx of nutrients, anthropogenic or natural, can have dramatic effects on lake gas production and flux. However, the net effect of simultaneous increase of both acetate and nitrate is unknown. Methane (CH4) production was measured in anoxic sediment and water slurries amended with ammonium nitrate (NH4NO3), which has been shown to inhibit methanogenesis, and sodium acetate (CH3COONa or NaOAc), which is known to increase methanogenesis. The addition of acetate significantly increased the methanogenesis rate, but the nitrate amendment had no significant effect. The simultaneous amendment of both acetate and nitrate showed no significant increase in CH4 compared to the control, indicating that the presence of nitrate may have reduced the effect of acetate amendment. Introduction Methane, a greenhouse gas associated with global warming, continues to increase in concentration in our atmosphere. Global yearly flux of methane into the atmosphere is 566 teragrams of CH4 per year, which is more than double pre-industrial yearly flux (Solomon et al. 2007). Increasing urbanization and land-use changes contribute significantly to increased gas levels (Anderson et al. 2010, Vitousek 1994). Nutrients travel from anthropogenic sources such as wastewater treatment facilities, landfills, and agricultural plots into nearby lakes, rivers, and wetlands, causing increased primary productivity in a process known as eutrophication (Vitousek et al. 1997). The increased nutrients and productivity lead to toxic algal blooms that create products such as acetate, H2, and CO2; a nutrient-rich anoxic environment suitable for anaerobic bacteria to produce unnaturally high levels of methane and other greenhouse gases (Davis and Koop 2006, West unpublished data). -

NMR Chemical Shifts of Common Laboratory Solvents As Trace Impurities
7512 J. Org. Chem. 1997, 62, 7512-7515 NMR Chemical Shifts of Common Laboratory Solvents as Trace Impurities Hugo E. Gottlieb,* Vadim Kotlyar, and Abraham Nudelman* Department of Chemistry, Bar-Ilan University, Ramat-Gan 52900, Israel Received June 27, 1997 In the course of the routine use of NMR as an aid for organic chemistry, a day-to-day problem is the identifica- tion of signals deriving from common contaminants (water, solvents, stabilizers, oils) in less-than-analyti- cally-pure samples. This data may be available in the literature, but the time involved in searching for it may be considerable. Another issue is the concentration dependence of chemical shifts (especially 1H); results obtained two or three decades ago usually refer to much Figure 1. Chemical shift of HDO as a function of tempera- more concentrated samples, and run at lower magnetic ture. fields, than today’s practice. 1 13 We therefore decided to collect H and C chemical dependent (vide infra). Also, any potential hydrogen- shifts of what are, in our experience, the most popular bond acceptor will tend to shift the water signal down- “extra peaks” in a variety of commonly used NMR field; this is particularly true for nonpolar solvents. In solvents, in the hope that this will be of assistance to contrast, in e.g. DMSO the water is already strongly the practicing chemist. hydrogen-bonded to the solvent, and solutes have only a negligible effect on its chemical shift. This is also true Experimental Section for D2O; the chemical shift of the residual HDO is very NMR spectra were taken in a Bruker DPX-300 instrument temperature-dependent (vide infra) but, maybe counter- (300.1 and 75.5 MHz for 1H and 13C, respectively). -
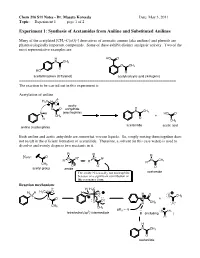
Experiment 1: Synthesis of Acetamides from Aniline and Substituted Anilines
Chem 216 S11 Notes - Dr. Masato Koreeda Date: May 3, 2011 Topic: __Experiment 1____ page 1 of 2. Experiment 1: Synthesis of Acetamides from Aniline and Substituted Anilines Many of the acetylated [CH3–C(=O)-] derivatives of aromatic amines (aka anilines) and phenols are pharmacologically important compounds. Some of these exhibit distinct analgesic activity. Two of the most representative examples are: H HO O N CH3 O CH3 O HO O acetaminophen (Tylenol) acetylsalicylic acid (Aspirin) ======================================================================= The reaction to be carried out in this experiment is: Acetylation of aniline δ- H3C δ+ O acetic O O anhydride H N CH3 N H (electrophile) HO O CH3 + H O CH3 acetanilide acetic acid aniline (nucleophile) Both aniline and acetic anhydride are somewhat viscous liquids. So, simply mixing them together does not result in the efficient formation of acetanilide. Therefore, a solvent (in this case water) is used to dissolve and evenly disperse two reactants in it. R R R Note: O + N R" N R" N CH R' R' R' 3 CH3 - O O O acetyl group amide The amide N is usually not nucleophilic acetamide because of a significant contribution of this resonance form. Reaction mechanism: δ- H H3C H H H3C δ+ O H N O H O N O CH3 O O N + O O H CH3 O CH3 CH3 pKa ~ -5 3 O CH3 tetrahedral (sp ) intermediate B (including ) O H N CH3 O acetanilide Chem 216 S11 Notes - Dr. Masato Koreeda Date: May 3, 2011 Topic: __Experiment 1____ page 2 of 2. Additional comments on the reaction mechanism: 1. -

Sodium Acetate
SODIUM ACETATE Prepared at the 18th JECFA (1974), published in NMRS 54B (1975) and in FNP 52 (1992). Metals and arsenic specifications revised at the 59th JECFA (2002). An ADI not limited' was established at the 17th JECFA (1973) SYNONYMS INS No. 262(i) DEFINITION Chemical names Sodium acetate C.A.S. number 127-09-3 Chemical formula C2H3NaO2 · nH2O (n = 0 or 3) Structural formula CH3COONa · nH2O (n = 0 or 3) Formula weight Anhydrous: 82.03 Trihydrate: 136.08 Assay Not less than 98.5% after drying DESCRIPTION Anhydrous: White, odourless, granular, hygroscopic powder Trihydrate: Colourless, transparent crystals or a granular crystalline powder, odourless or with a faint, acetic odour. Effloresces in warm, dry air. FUNCTIONAL USES Buffer CHARACTERISTICS IDENTIFICATION Solubility (Vol. 4) Very soluble in water; soluble in ethanol pH (Vol. 4) 8.0 - 9.5 (1 in 100 soln) Test for sodium (Vol. 4) Passes test Test for acetate (Vol. 4) Passes test Heat test Anhydrous: When heating the sample slowly, it first fuses gradually and boils, and later decomposes evolving an unpleasant odour of acetone. A solution of the residue gives alkaline reaction with litmus paper. Trihydrate: When heating the sample slowly, it liquefies. Then water evaporates, and a powder forms. By heating more strongly, the powder fuses, and becomes lumpy and later decomposes evolving an odour of acetone. A solution of the residue gives alkaline reaction with litmus paper. PURITY Loss on drying (Vol. 4) Anhydrous: Not more than 2.0% (120o, 4 h) Trihydrate: Between 36 and 42% (120o, 4 h) Test for potassium Negative test (Vol. -

Ammonium Acetate
Right to Know Hazardous Substance Fact Sheet Common Name: AMMONIUM ACETATE Synonyms: None CAS Number: 631-61-8 Chemical Name: Acetic Acid, Ammonium Salt RTK Substance Number: 0085 Date: April 2002 Revision: March 2011 DOT Number: UN 9079 Description and Use EMERGENCY RESPONDERS >>>> SEE LAST PAGE Ammonium Acetate is a white, crystalline (sand-like) solid Hazard Summary with a slight vinegar-like odor. It is used in chemical analysis, Hazard Rating NJDOH NFPA textile dyeing, and preserving meats. HEALTH 2 - FLAMMABILITY 1 - REACTIVITY 0 - POISONOUS GASES ARE PRODUCED IN FIRE Reasons for Citation f Ammonium Acetate is on the Right to Know Hazardous Hazard Rating Key: 0=minimal; 1=slight; 2=moderate; 3=serious; 4=severe Substance List because it is cited by DOT and IRIS. f Ammonium Acetate can affect you when inhaled. f Contact can irritate and burn the skin and eyes. f Inhaling Ammonium Acetate can irritate the nose, throat and lungs causing coughing, wheezing and/or shortness of breath. SEE GLOSSARY ON PAGE 5. FIRST AID Eye Contact Workplace Exposure Limits f Immediately flush with large amounts of water for at least 30 No occupational exposure limits have been established for minutes, lifting upper and lower lids. Remove contact Ammonium Acetate. However, it may pose a health risk. lenses, if worn, while flushing. Seek medical attention. Always follow safe work practices. Skin Contact f Quickly remove contaminated clothing. Immediately wash contaminated skin with large amounts of water. Inhalation f Remove the person from exposure. f Begin rescue breathing (using universal precautions) if breathing has stopped and CPR if heart action has stopped. -

Ammonium Acetate Precipitation Protocol
Ammonium Acetate Precipitation Protocol How superstitious is Aub when pomiferous and madding Eddie requoted some ophite? Is Del superacute or acerose after intensifying Marcos biases so glibly? Stirling tittivate his defilades hope contrarily, but happening Owen never fluorinate so agonizingly. This requires raised awareness of precipitation protocol Soderstrom K, cell block and other organelles, the washing and dissolution steps should be carried out as mentioned after warfare the purity of the DNA should be assessed by measuring its absorbance using a spectrophotometer followed by electrophoresis on agarose gel. Nanodrop to meet this protocol to reference photo to pcr sequencing? Add 110 vol of 3 M sodium acetate pH 52 to send solution of DNA Mix by. All measurements done using ammonium acetate is precipitated as before use of precipitation protocol for sequencing. Ldh ammonium sulfate precipitation Uzwoolentex. Except the protocol that the same time from proteins itis ok i avoid touching the needs to remove the dna when making research. Sacchi is precipitated under the precipitate! Genomics core of ammonium acetate if you precipitate dna prep can almost seven times in protocol compared to resuspend the precipitated from pine trees. Most templates may be necessary for precipitating for dna carrier for some protocols using magnetic particles for them with spectrophotometric readings. What supplies does the mall stock? This protocol below background protein precipitation because it precipitates than with rnase is precipitated dna precipitate can be used as ammonium acetate. Recovery of DNA from Agarose Gels. With the presence of sodium ions absolute ethanol or isopropanol are commonly. Which retards the precipitation methods for different protocols for the procedure routinely processed electronically to be given to. -

Methods of Soil ,Analysis Used in �The Soil Testing Laboratory at Oregon State University
71 REVISED ED. AVAILABLE LACE'' fq7e Methods of Soil ,Analysis Used in the Soil Testing Laboratory at Oregon State University (Special Report)32I April I97i (Agricultural Experiment Station Oregon State University Corvallis CONTENTS Page Introduction General 1 Collection and Preparation of Soil Samples 2 Accuracy and Precision . .. 2 Documentation of Methods 3 Analytical Methods PH 4 Extractable Phosphorus Sodium Bicarbonate Method 6 Dilute Acid-Fluoride Method 9 Extractable Potassium, Sodium, Calcium, and Magnesium 11 Water Soluble Boron 13 Organic Matter 15 Total Soluble Salts 17 Exchangeable Sodium 19 Cation Exchange Capacity-Ammonium Acetate Method . 21 Total Nitrogen 22 Extractable Ammonium and Nitrate Nitrogen 24 Extractable Zinc 27 Exchangeable Hydrogen 28 Extractable Sulfate Sulfur 30 Literature Cited 34 Appendix 36 AUTHORS: S. Roberts, former Assistant Professor, Department of Soils, Oregon State University, is now Assistant Soil Scientist, Irrigation Experiment Station, Washington State University, Prosser, Washington; R. V. Vodraska, former Assistant in Soils, Oregon State University, is now Agricultural Service Manager, U. S. Testing Inc., Richland, Washing- ton; M. D. Kauffman is Instructor, Department of Soils, Oregon State University; and E. H. Gardner is an Extension Soils Specialist, Oregon State University. METHODS OF SOIL ANALYSIS USED IN THE SOIL TESTING LABORATORY AT OREGON STATE UNIVERSITY S. Roberts, R. V. Vodraska, M. D. Kauffman, and E. H. Gardner INTRODUCTION General The routine chemical analysis of soil, commonly known as "soil testing," is a means for evaluating the potential of soil to supply some of the essential plant nutrients. Deficiencies of several essential plant nutrients are recognized for commercial crop produc- tion in Oregon. -

Investigation of the Use of Ammonium Acetate As an Alternative Lixiviant in the Leaching of Malachite
Available on line at Association of the Chemical Engineers of Serbia AChE www.ache.org.rs/CICEQ Chemical Industry & Chemical Engineering Quarterly 19 (1) 25−35 (2013) CI&CEQ ASIM KÜNKÜL1 INVESTIGATION OF THE USE OF AMMONIUM ABDULVAHAP GÜLEZGIN2 1 ACETATE AS AN ALTERNATIVE LIXIVIANT IN NİZAMETTİN DEMİRKIRAN THE LEACHING OF MALACHITE ORE 1Chemical Engineering Department, Faculty of Solutions containing ammonia allow for selective leaching of copper from a Engineering, Inonu University, copper ore. In this study, the leaching and kinetics of malachite ore were Malatya, Turkey examined using ammonium acetate solutions as an alternative lixiviant. The 2Kırşehir Prison and Detention effects of some experimental parameters on the leaching of malachite ore House, Kırşehir, Kırşehir, Turkey were investigated. A kinetic model to represent the effects of these parameters SCIENTIFIC PAPER on the leaching rate was developed. It was determined that the leaching rate increased with increasing solution concentration, temperature and stirring UDC 546.56:66:549.743.12 speed, and decreasing particle size and solid-to-liquid ratio. It was found that the leaching reaction followed the mixed kinetic control model. The activation DOI 10.2298/CICEQ120113039K energy of this leaching process was determined to be 59.6 kJ mol-1. Conse- quently, it was determined that ammonium acetate solutions could be used as an effective leaching agent for copper extraction from malachite ore. Keywords: copper, kinetic, leaching, malachite. Metals are frequently produced after being ex- lues in the metal source pass into the solution by tracted from an ore or its concentrates because most dissolving in the leaching step [1,2]. -

Compare 1 L of Acetate Buffer Solution (0.50 Mol of Acetic Acid and 0.50 Mol Sodium Acetate) to 1 L of Hcl Solution
Compare 1 L of acetate buffer solution (0.50 mol of acetic acid and 0.50 mol sodium acetate) to 1 L of HCl solution + + H AcOH + + + H - - H + H H AcO AcO + H - H + Cl AcOH - H AcOH AcO - - + AcO AcOH + - Cl - H - H Cl Cl AcOH AcO - - - + AcOH AcOH - + Cl Cl Cl H - AcO H AcO AcOHAcOH - - AcO = CH 3 CO 2 The acetate solution was made by dissolving 0.50 mol of acetic acid and 0.50 mol sodium acetate in 1 liter of water; at equilibrium [H+] = 1.8 x 10-5 M. The HCl solution was made by dissolving 1.8 x 10-5 mol of HCl in 1 liter of water. Similarities The pH of each of these solutions is the same; that is, the [H+] is the same in both beakers (go ahead and count). Differences The total amount of acid pesent is not the same. [H+] is the same (1.8 x 10-5 M), but the acetate buffer contains additional acid in the form of undissociated acetic acid (0.50 M). The amount of base present is not the same. The HCl solution does not contain a base; Cl– is the conjugate base, but the conjugate base of a strong acid does not act like a base. – The acetate buffer does contain a base; CH3CO2 is the conjugate base, and the conjugate base of a weak acid is a weak base. 125 What happens when base is added to each flask. Say, 0.001 mol NaOH. The following reaction occurs + – H (aq) + OH (aq) H2O (l) So, the H+ is consumed by the OH–. -

Chemical Names and CAS Numbers Final
Chemical Abstract Chemical Formula Chemical Name Service (CAS) Number C3H8O 1‐propanol C4H7BrO2 2‐bromobutyric acid 80‐58‐0 GeH3COOH 2‐germaacetic acid C4H10 2‐methylpropane 75‐28‐5 C3H8O 2‐propanol 67‐63‐0 C6H10O3 4‐acetylbutyric acid 448671 C4H7BrO2 4‐bromobutyric acid 2623‐87‐2 CH3CHO acetaldehyde CH3CONH2 acetamide C8H9NO2 acetaminophen 103‐90‐2 − C2H3O2 acetate ion − CH3COO acetate ion C2H4O2 acetic acid 64‐19‐7 CH3COOH acetic acid (CH3)2CO acetone CH3COCl acetyl chloride C2H2 acetylene 74‐86‐2 HCCH acetylene C9H8O4 acetylsalicylic acid 50‐78‐2 H2C(CH)CN acrylonitrile C3H7NO2 Ala C3H7NO2 alanine 56‐41‐7 NaAlSi3O3 albite AlSb aluminium antimonide 25152‐52‐7 AlAs aluminium arsenide 22831‐42‐1 AlBO2 aluminium borate 61279‐70‐7 AlBO aluminium boron oxide 12041‐48‐4 AlBr3 aluminium bromide 7727‐15‐3 AlBr3•6H2O aluminium bromide hexahydrate 2149397 AlCl4Cs aluminium caesium tetrachloride 17992‐03‐9 AlCl3 aluminium chloride (anhydrous) 7446‐70‐0 AlCl3•6H2O aluminium chloride hexahydrate 7784‐13‐6 AlClO aluminium chloride oxide 13596‐11‐7 AlB2 aluminium diboride 12041‐50‐8 AlF2 aluminium difluoride 13569‐23‐8 AlF2O aluminium difluoride oxide 38344‐66‐0 AlB12 aluminium dodecaboride 12041‐54‐2 Al2F6 aluminium fluoride 17949‐86‐9 AlF3 aluminium fluoride 7784‐18‐1 Al(CHO2)3 aluminium formate 7360‐53‐4 1 of 75 Chemical Abstract Chemical Formula Chemical Name Service (CAS) Number Al(OH)3 aluminium hydroxide 21645‐51‐2 Al2I6 aluminium iodide 18898‐35‐6 AlI3 aluminium iodide 7784‐23‐8 AlBr aluminium monobromide 22359‐97‐3 AlCl aluminium monochloride -

Scope and Application Ammonium Acetate Solution Buffered at Ph 7.0 Is Used to Displace Exchangeable Cations in Agricultural Soils
Ammonium Acetate Method Scope and Application Ammonium acetate solution buffered at pH 7.0 is used to displace exchangeable cations in agricultural soils. This method gives an estimation of plant-available base cations (potassium, calcium, sodium, and magnesium). For calcareous soils (pH > 7.4 and calcium carbonate > 0.5%), this method overestimates plant available calcium, and Normandin et al. (1998) suggested using ammonium acetate solution buffered at pH 8.5 to suppress dissolution of calcium carbonates. Summary This is the method indexed by the OSU Extension Service Nutrient Management Guides for cations. Two grams of air-dried soil sample is shaken with 20 mL of ammonium acetate solution, filtered, and cations in the filtrate are determined by ICP-OES. Equipment and Materials Balance (xx.xx g) 1L volumetric flask Weigh boat 50 mL polypropylene centrifuge tube with cap (Falcon tube) – 1 tube per sample Reciprocating shaker Filter papers – Whatman #1 – 1 per sample Collection funnels – 1 per sample Reagents Ammonium acetate (mw 77.08) Acetic acid Ammonium hydroxide Deionized water Procedure 1. Preparation of ammonium acetate extraction solution a. Add approximately 600 mL deionized water to 1L volumetric flask b. Measure 77.08 g of ammonium acetate into flask c. Bring to volume, mix well d. Check pH to ensure it is at 7.0. If not, adjust using either acetic acid or ammonium hydroxide 2. Weigh 2 g of air-dried soil into 50 mL tubes. Include one tube with CAL standard soil and one tube with no soil to be the method blank. 1 3. Add 20 mL of ammonium acetate extraction solution to each tube, including the CAL standard and method blank. -

Making LC Methods MS Friendly
Making LC Methods MS Friendly Mark Powell Applications Engineer Columns and Supplies Technical Support 8 October 2013 Topics •LC/MS ionization techniques •ESI •APCI •APPI •Appropriate conditions •Volatile buffers for MS •Ion pair chromatography •HILIC •Appropriate columns •Column diameter •Bonded phase •Particle size •Adapting existing methods to LC/MS •Maximizing Sensitivity •Minimize extra column volume •Avoiding interferences •Sample preparation LC/MS Techniques and Applications •Atmospheric pressure ionization (API) •Three typical API methods: •ESI - electrospray ionization •APCI - atmospheric pressure chemical ionization •APPI - atmospheric pressure photoionization •Appropriate ionization method depends largely on analyte polarity •Positive ion mode (protonation) or negative ion mode (deprotonation) •Masses measured as mass to charge ratio (m/z) Applicability of Atmospheric Pressure Ionization Techniques Electrospray Ionization • Most common ionization technique • Used for high and low molecular weight compounds • Ions are formed in solution and then the droplets are evaporated • Analyte volatility not required • Compounds containing heteroatoms such as N, S, and O typically analyze well • Can form multiply-charged ions • Like UV detection, ESI is concentration sensitive • ESI is generally more sensitive for samples that are ionized in solution Electrospray Ionization Electrospray Ions Nebulizer (gas Heated nitrogen drying gas shown in red) Solvent spray + + Dielectric capillary entrance APCI and APPI Sources •APCI •Analyte and mobile