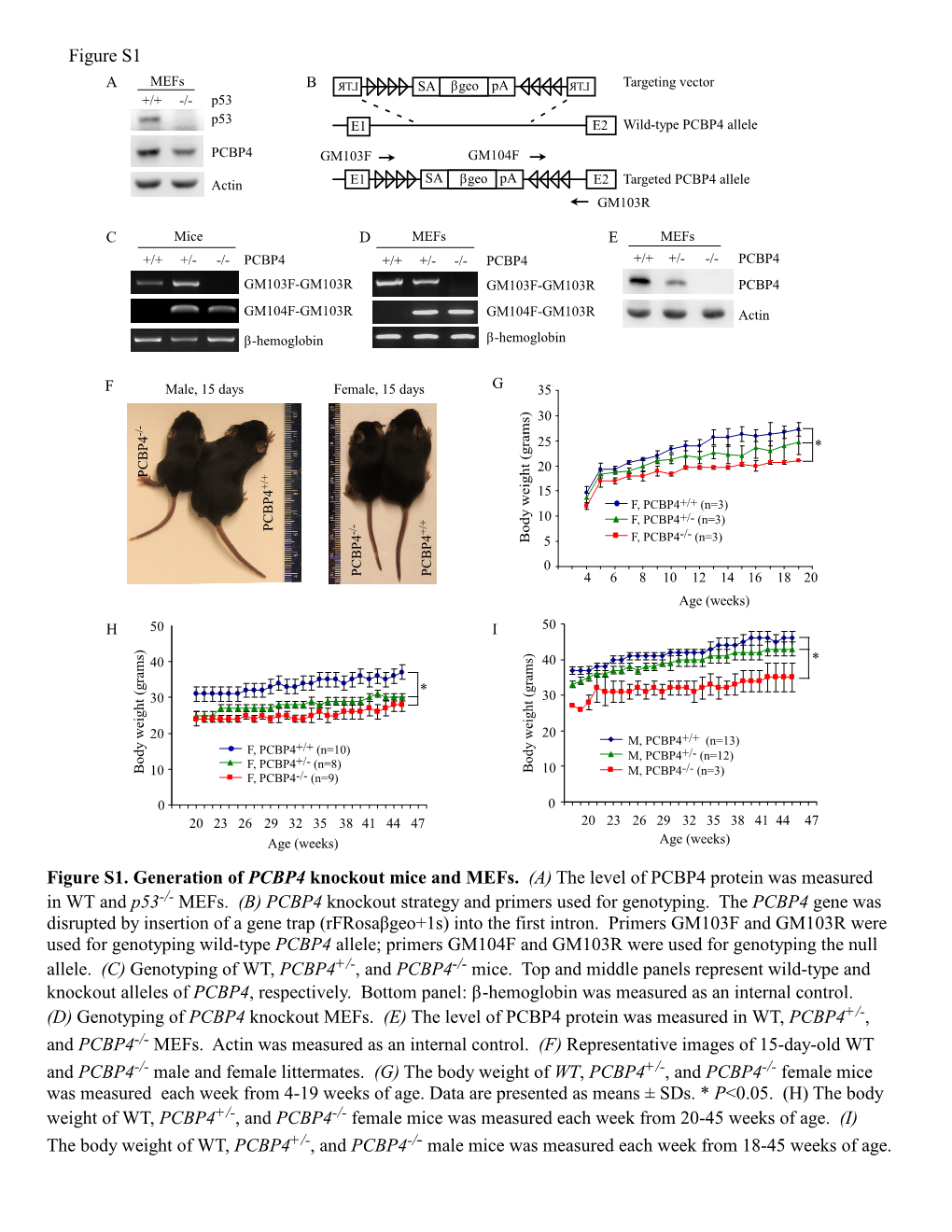
(A) the Level of PCBP4 Protein Was Measured in WT and P53-/- Mefs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MCG10 (PCBP4) (NM 033008) Human Recombinant Protein Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TP300749 MCG10 (PCBP4) (NM_033008) Human Recombinant Protein Product data: Product Type: Recombinant Proteins Description: Recombinant protein of human poly(rC) binding protein 4 (PCBP4), transcript variant 3 Species: Human Expression Host: HEK293T Tag: C-Myc/DDK Predicted MW: 41.3 kDa Concentration: >50 ug/mL as determined by microplate BCA method Purity: > 80% as determined by SDS-PAGE and Coomassie blue staining Buffer: 25 mM Tris.HCl, pH 7.3, 100 mM glycine, 10% glycerol Preparation: Recombinant protein was captured through anti-DDK affinity column followed by conventional chromatography steps. Storage: Store at -80°C. Stability: Stable for 12 months from the date of receipt of the product under proper storage and handling conditions. Avoid repeated freeze-thaw cycles. RefSeq: NP_127501 Locus ID: 57060 UniProt ID: P57723, A0A024R2Y0 RefSeq Size: 2040 Cytogenetics: 3p21.2 RefSeq ORF: 1209 Synonyms: CBP; LIP4; MCG10 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 MCG10 (PCBP4) (NM_033008) Human Recombinant Protein – TP300749 Summary: This gene encodes a member of the KH-domain protein subfamily. Proteins of this subfamily, also referred to as alpha-CPs, bind to RNA with a specificity for C-rich pyrimidine regions. Alpha-CPs play important roles in post-transcriptional activities and have different cellular distributions. -

Spermatozoal Gene KO Studies for the Highly Present Paternal Transcripts
Spermatozoal gene KO studies for the highly present paternal Potential maternal interactions Conclusions of the KO studies on the maternal gene transcripts detected by GeneMANIA candidates for interaction with the paternal Fbxo2 Selective cochlear degeneration in mice lacking Cul1, Fbxl2, Fbxl3, Fbxl5, Cul-1 KO causes early embryonic lethality at E6.5 before Fbxo2 Fbxo34, Fbxo5, Itgb1, Rbx1, the onset of gastrulation http://www.jneurosci.org/content/27/19/5163.full Skp1a http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3641602/. Loss of Cul1 results in early embryonic lethality and Another KO study showed the following: Loss of dysregulation of cyclin E F-box only protein 2 (Fbxo2) disrupts levels and http://www.ncbi.nlm.nih.gov/pubmed/10508527?dopt=Abs localization of select NMDA receptor subunits, tract. and promotes aberrant synaptic connectivity http://www.ncbi.nlm.nih.gov/pubmed/25878288. Fbxo5 (or Emi1): Regulates early mitosis. KO studies shown lethal defects in preimplantation embryo A third KO study showed that Fbxo2 regulates development http://mcb.asm.org/content/26/14/5373.full. amyloid precursor protein levels and processing http://www.jbc.org/content/289/10/7038.long#fn- Rbx1/Roc1: Rbx1 disruption results in early embryonic 1. lethality due to proliferation failure http://www.pnas.org/content/106/15/6203.full.pdf & Fbxo2 has been found to be involved in http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2732615/. neurons, but it has not been tested for potential decreased fertilization or pregnancy rates. It Skp1a: In vivo interference with Skp1 function leads to shows high expression levels in testis, indicating genetic instability and neoplastic transformation potential other not investigated functions http://www.ncbi.nlm.nih.gov/pubmed/12417738?dopt=Abs http://biogps.org/#goto=genereport&id=230904 tract. -

Potential Sperm Contributions to the Murine Zygote Predicted by in Silico Analysis
REPRODUCTIONRESEARCH Potential sperm contributions to the murine zygote predicted by in silico analysis Panagiotis Ntostis1,2, Deborah Carter3, David Iles1, John Huntriss1, Maria Tzetis2 and David Miller1 1Leeds Institute of Cardiovascular and Metabolic Medicine, University of Leeds, Leeds, West Yorkshire, UK, 2Department of Medical Genetics, St. Sophia’s Children Hospital, School of Medicine, National and Kapodistrian University of Athens, Athens, Attiki, Greece and 3Leeds Institute of Molecular Medicine, University of Leeds, Leeds, West Yorkshire, UK Correspondence should be addressed to D Miller; Email: [email protected] Abstract Paternal contributions to the zygote are thought to extend beyond delivery of the genome and paternal RNAs have been linked to epigenetic transgenerational inheritance in different species. In addition, sperm–egg fusion activates several downstream processes that contribute to zygote formation, including PLC zeta-mediated egg activation and maternal RNA clearance. Since a third of the preimplantation developmental period in the mouse occurs prior to the first cleavage stage, there is ample time for paternal RNAs or their encoded proteins potentially to interact and participate in early zygotic activities. To investigate this possibility, a bespoke next-generation RNA sequencing pipeline was employed for the first time to characterise and compare transcripts obtained from isolated murine sperm, MII eggs and pre-cleavage stage zygotes. Gene network analysis was then employed to identify potential interactions between paternally and maternally derived factors during the murine egg-to-zygote transition involving RNA clearance, protein clearance and post-transcriptional regulation of gene expression. Our in silico approach looked for factors in sperm, eggs and zygotes that could potentially interact co-operatively and synergisticallyp during zygote formation. -

A Causal Gene Network with Genetic Variations Incorporating Biological Knowledge and Latent Variables
A CAUSAL GENE NETWORK WITH GENETIC VARIATIONS INCORPORATING BIOLOGICAL KNOWLEDGE AND LATENT VARIABLES By Jee Young Moon A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Statistics) at the UNIVERSITY OF WISCONSIN–MADISON 2013 Date of final oral examination: 12/21/2012 The dissertation is approved by the following members of the Final Oral Committee: Brian S. Yandell. Professor, Statistics, Horticulture Alan D. Attie. Professor, Biochemistry Karl W. Broman. Professor, Biostatistics and Medical Informatics Christina Kendziorski. Associate Professor, Biostatistics and Medical Informatics Sushmita Roy. Assistant Professor, Biostatistics and Medical Informatics, Computer Science, Systems Biology in Wisconsin Institute of Discovery (WID) i To my parents and brother, ii ACKNOWLEDGMENTS I greatly appreciate my adviser, Prof. Brian S. Yandell, who has always encouraged, inspired and supported me. I am grateful to him for introducing me to the exciting research areas of statis- tical genetics and causal gene network analysis. He also allowed me to explore various statistical and biological problems on my own and guided me to see the problems in a bigger picture. Most importantly, he waited patiently as I progressed at my own pace. I would also like to thank Dr. Elias Chaibub Neto and Prof. Xinwei Deng who my adviser arranged for me to work together. These three improved my rigorous writing and thinking a lot when we prepared the second chapter of this dissertation for publication. It was such a nice opportunity for me to join the group of Prof. Alan D. Attie, Dr. Mark P. Keller, Prof. Karl W. Broman and Prof. -

Mice Deficient in Poly(C)-Binding Protein 4 Are Susceptible to Spontaneous Tumors Through Increased Expression of ZFP871 That Targets P53 for Degradation
Downloaded from genesdev.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Mice deficient in poly(C)-binding protein 4 are susceptible to spontaneous tumors through increased expression of ZFP871 that targets p53 for degradation Wensheng Yan,1 Ariane Scoumanne,1 Yong-Sam Jung, Enshun Xu, Jin Zhang, Yanhong Zhang, Cong Ren, Pei Sun, and Xinbin Chen Comparative Oncology Laboratory, School of Veterinary Medicine, School of Medicine, University of California at Davis, Davis, California 95616, USA Poly(C)-binding protein 4 (PCBP4), also called MCG10 and a target of p53, plays a role in the cell cycle and is im- plicated in lung tumor suppression. Here, we found that PCBP4-deficient mice are prone to lung adenocarcinoma, lymphoma, and kidney tumor and that PCBP4-deficient mouse embryo fibroblasts (MEFs) exhibit enhanced cell proliferation but decreased cellular senescence. We also found that p53 expression is markedly reduced in PCBP4- deficient MEFs and mouse tissues, suggesting that PCBP4 in turn regulates p53 expression. To determine how PCBP4 regulates p53 expression, PCBP4 targets were identified by RNA immunoprecipitation followed by RNA sequencing (RNA-seq). We found that the transcript encoding ZFP871 (zinc finger protein 871; also called ZNF709 in humans) interacts with and is regulated by PCBP4 via mRNA stability. Additionally, we found that ZFP871 phys- ically interacts with p53 and MDM2 proteins. Consistently, ectopic expression of ZFP871 decreases—whereas knockdown of ZFP871 increases—p53 protein stability through a proteasome-dependent degradation pathway. Moreover, loss of ZFP871 reverses the reduction of p53 expression by lack of PCBP4, and thus increased expression of ZFP871 is responsible for decreased expression of p53 in the PCBP4-deficient MEFs and mouse tissues. -

Anti-PCBP4 Antibody (ARG64279)
Product datasheet [email protected] ARG64279 Package: 100 μg anti-PCBP4 antibody Store at: -20°C Summary Product Description Goat Polyclonal antibody recognizes PCBP4 Tested Reactivity Hu Predict Reactivity Ms, Rat, Dog Tested Application WB Specificity This antibody is expected to recognise all reported isoforms (NP_065151.1, NP_127501.1, NP_127502.1 and NP_127503.1). Host Goat Clonality Polyclonal Isotype IgG Target Name PCBP4 Antigen Species Human Immunogen C-NGSKKAERQKFSPY Conjugation Un-conjugated Alternate Names Alpha-CP4; Poly; CBP; LIP4; rC; MCG10 Application Instructions Application table Application Dilution WB 1 - 3 µg/ml Application Note WB: Recommend incubate at RT for 1h. * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Calculated Mw 41 kDa Properties Form Liquid Purification Purified from goat serum by ammonium sulphate precipitation followed by antigen affinity chromatography using the immunizing peptide. Buffer Tris saline (pH 7.3), 0.02% Sodium azide and 0.5% BSA Preservative 0.02% Sodium azide Stabilizer 0.5% BSA Concentration 0.5 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated www.arigobio.com 1/2 freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Database links GeneID: 57060 Human Swiss-port # P57723 Human Background This gene encodes a member of the KH-domain protein subfamily. -

CREB-Dependent Transcription in Astrocytes: Signalling Pathways, Gene Profiles and Neuroprotective Role in Brain Injury
CREB-dependent transcription in astrocytes: signalling pathways, gene profiles and neuroprotective role in brain injury. Tesis doctoral Luis Pardo Fernández Bellaterra, Septiembre 2015 Instituto de Neurociencias Departamento de Bioquímica i Biologia Molecular Unidad de Bioquímica y Biologia Molecular Facultad de Medicina CREB-dependent transcription in astrocytes: signalling pathways, gene profiles and neuroprotective role in brain injury. Memoria del trabajo experimental para optar al grado de doctor, correspondiente al Programa de Doctorado en Neurociencias del Instituto de Neurociencias de la Universidad Autónoma de Barcelona, llevado a cabo por Luis Pardo Fernández bajo la dirección de la Dra. Elena Galea Rodríguez de Velasco y la Dra. Roser Masgrau Juanola, en el Instituto de Neurociencias de la Universidad Autónoma de Barcelona. Doctorando Directoras de tesis Luis Pardo Fernández Dra. Elena Galea Dra. Roser Masgrau In memoriam María Dolores Álvarez Durán Abuela, eres la culpable de que haya decidido recorrer el camino de la ciencia. Que estas líneas ayuden a conservar tu recuerdo. A mis padres y hermanos, A Meri INDEX I Summary 1 II Introduction 3 1 Astrocytes: physiology and pathology 5 1.1 Anatomical organization 6 1.2 Origins and heterogeneity 6 1.3 Astrocyte functions 8 1.3.1 Developmental functions 8 1.3.2 Neurovascular functions 9 1.3.3 Metabolic support 11 1.3.4 Homeostatic functions 13 1.3.5 Antioxidant functions 15 1.3.6 Signalling functions 15 1.4 Astrocytes in brain pathology 20 1.5 Reactive astrogliosis 22 2 The transcription -

Cancer Theâ•Ÿrbpâ•Žeutics-RNA-Binding Proteins As
Pharmacology & Therapeutics 203 (2019) 107390 Contents lists available at ScienceDirect Pharmacology & Therapeutics journal homepage: www.elsevier.com/locate/pharmthera Cancer the‘RBP’eutics–RNA-binding proteins as therapeutic targets for cancer Shakur Mohibi, Xinbin Chen, Jin Zhang ⁎ Comparative Oncology Laboratory, Schools of Veterinary Medicine and Medicine, University of California at Davis, United States article info abstract Available online 11 July 2019 RNA-binding proteins (RBPs) play a critical role in the regulation of various RNA processes, including splicing, cleavage and polyadenylation, transport, translation and degradation of coding RNAs, non-coding RNAs and Keywords: microRNAs. Recent studies indicate that RBPs not only play an instrumental role in normal cellular processes RNA-binding proteins but have also emerged as major players in the development and spread of cancer. Herein, we review the current RNA-binding domains knowledge about RNA binding proteins and their role in tumorigenesis as well as the potential to target RBPs for Cancer therapeutics cancer therapeutics. Small molecules © 2019 Elsevier Inc. All rights reserved. siRNA Peptide Aptamer Contents 1. Introduction................................................ 1 2. RNA-bindingdomains........................................... 2 3. AberrantexpressionofRBPsincancer.................................... 4 4. RBPsastherapeutictargetsforcancer.................................... 9 5. Conclusionsandfutureperspectives.................................... 14 DeclarationsofCompetingInterest...................................... -

Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name poly(rC) binding protein 4 Gene Symbol Pcbp4 Organism Mouse Gene Summary Description Not Available Gene Aliases 1200003L19Rik, AlphaCP-4 RefSeq Accession No. NC_000075.6, NT_039477.8 UniGene ID Mm.286394 Ensembl Gene ID ENSMUSG00000023495 Entrez Gene ID 59092 Assay Information Unique Assay ID qMmuCIP0028916 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENSMUST00000024260 Amplicon Context Sequence TCCGCGCCTGTCAAACCCCTCATTGTTCGCAGCTGATGTCACTCGCAGTTGTGA GCGGCCGCCTTTCCCGGGGACAATGTGGGACTGAGCGGCCCAGCCGCTGCCG CCGCCGCAGAACAGCCCCAGCGAGAGCTAGCTGAGCCACAGTGTGGACACACT CCCAGGCCGCTGCGGCCCCAGCCCCGCCTGACAGGATGAGCAGTTCAGATGCG GGGCTG Amplicon Length (bp) 188 Chromosome Location 9:106453878-106460092 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 97 R2 0.9998 cDNA Cq 25.32 cDNA Tm (Celsius) 91.5 gDNA Cq 33.15 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Pcbp4, Mouse Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Mouse Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

High Rate of Molecular Alteration in Histologically Tumour-Free Bronchial Epithelium of NSCLC Patients Detected by Multicolour Fluorescence in Situ Hybridisation
1233-1240 28/3/06 14:14 Page 1233 ONCOLOGY REPORTS 15: 1233-1240, 2006 High rate of molecular alteration in histologically tumour-free bronchial epithelium of NSCLC patients detected by multicolour fluorescence in situ hybridisation WOLFGANG HILBE1, JUTTA AUBERGER1, STEPHAN DIRNHOFER5, THOMAS SCHMID3, MARTIN ERDEL2 and HANS-CHRISTOPH DUBA2,4 Departments of 1Internal Medicine, 2Medical Biology and Human Genetics, and 3Surgery, Medical University Innsbruck, Innsbruck; 4Department of Human Genetics, General Women and Children Hospital, Linz, Austria; 5Department of Pathology, University of Basel, Switzerland Received January 18, 2006; Accepted February 7, 2006 Abstract. Detection of molecular abnormalities could Introduction provide an essential tool for the diagnosis of non-small cell lung cancer (NSCLC) and defining patients at risk for early Non-small cell lung cancer (NSCLC) is the leading cause of relapse. Fluorescence in situ hybridisation (FISH) targeting cancer-related deaths in the Western world. The failure of 17 gene loci was applied to determine the frequency of antismoking campaigns, limitations in cytostatic treatment, molecular alteration in NSCLC probes and adjacent tumour- as well as the lack of efficient screening tests resulting in free bronchial epithelium. FISH was performed on fresh frozen delayed diagnoses are the reasons that patient outcome has specimens from 76 patients with histologically confirmed not improved substantially in recent years. New developments NSCLC and 54 specimens of adjacent tumour-free tissue. in the field of molecular biology have raised hopes for Routine autopsy lung tissue probes from 7 cancer-free patients improving the patient's clinical course. Molecular typing of served as a control group. Locus-specific (3p14.2, 3p21.2, bronchial epithelium could provide an essential tool for 3p21.3, 3p25.3, 5p15.2, 7p12, 8q24.12, 9p21, 13q14, and screening programs and, thus, early diagnosis (1-3). -

Quantitative Proteomics Analysis of Glioblastoma Cell Lines After Lncrna HULC Silencing Shan Ye1, Jing Wu2,3, Yiran Wang1, Yuchen Hu1, Tiantian Yin1 & Jie He1,2,3*
www.nature.com/scientificreports OPEN Quantitative proteomics analysis of glioblastoma cell lines after lncRNA HULC silencing Shan Ye1, Jing Wu2,3, Yiran Wang1, Yuchen Hu1, Tiantian Yin1 & Jie He1,2,3* Glioblastoma multiforme (GBM) is a life-threatening brain tumor. This study aimed to identify potential targets of the long noncoding RNA (lncRNA) HULC that promoted the progression of GBM. Two U87 cell lines were constructed: HULC-siRNA and negative control (NC). Quantitative real-time PCR (qRT-PCR) was performed to validate the transfection efciency of HULC silencing vector. Mass spectrometry (MS) was used to generate proteomic profles for the two cell lines. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to distinguish HULC-related genes and pathway mapping. Colony formation, Transwell, and wound- healing assays were used to investigate the functional efects of HULC knockdown on GBM. We identifed 112 up-regulated proteins and 24 down-regulated proteins from a total of 4360 quantifed proteins. GO enrichment illustrated that these proteins were mainly involved in organelle structure, catalysis, cell movement, and material metabolism. KEGG pathway analysis indicated that some of these proteins were signifcantly enriched in tight junction, metabolic pathways, and arachidonic acid metabolism. In vitro experiments demonstrated that HULC knockdown inhibited GBM cell proliferation, invasion, and migration. Our KEGG analyses revealed that PLA2G4A was a shared protein in several enriched pathways. HULC silencing signifcantly down-regulated the expression of PLA2G4A. Knockdown of HULC changed the proteomic characteristics of GBM and altered the behaviors of GBM cells. Specifcally, we identifed PLA2G4A as an HULC target in GBM. -

Modulation of the P53 Family Network by RNA-Binding Proteins
Review Article Modulation of the p53 family network by RNA-binding proteins Chris Lucchesi, Jin Zhang, Xinbin Chen Comparative Oncology Laboratory, School of Veterinary Medicine, School of Medicine, University of California at Davis, Davis, California 95616, USA Contributions: (I) Conception and design: All authors; (II) Administrative support: None; (III) Provision of study materials or patients: None; (IV) Collection and assembly of data: None; (V) Data analysis and interpretation: None; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Xinbin Chen. Comparative Oncology Laboratory, School of Veterinary Medicine, University of California, Davis, 2128A Tupper Hall, Davis, CA 95616, USA. Email: [email protected]. Abstract: Since its discovery more than three decades ago, tumor suppressor p53 has been shown to play pivotal roles in both maintaining genomic integrity and tumor suppression. p53 functions as a transcription factor responding to a multitude of cellular stressors, regulating the transcription of many genes involved in cell-cycle arrest, senescence, autophagy, and apoptosis. Extensive work has revealed that p53 is one of the most commonly mutated tumor suppressor genes. The last three decades have demonstrated that p53 activity is controlled through transcriptional regulation and posttranslational modifications. However, evolving work is now uncovering that p53, and other p53 family members, are post-transcriptionally regulated by multiple RNA-binding proteins (RBPs). Understanding the regulation of p53 by RBPs may potentially open up the possibility for cancer therapeutic intervention. This review focuses on the posttranscriptional regulation of p53, and p53 family members, by RNA binding proteins and the reciprocal feedback pathways between several RNA-biding proteins modulating p53, and p53 family members.