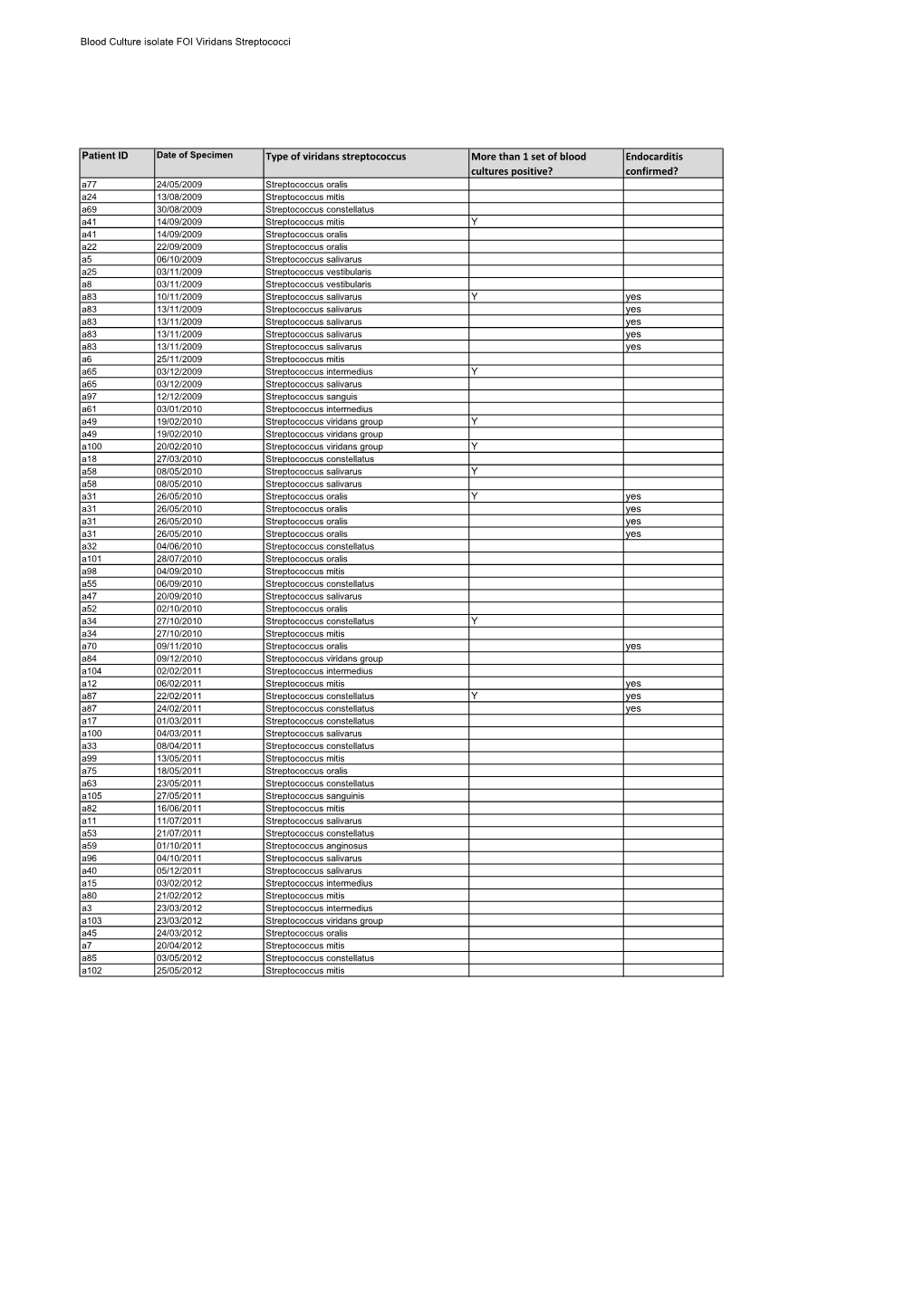
Type of Viridans Streptococcus More Than 1 Set of Blood Cultures Positive
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fecal Bacteriology of Colonic Polyp Patients and Control Patients'
[CANCER RESEARCH35,3407-3417,November1975] Fecal Bacteriology of Colonic Polyp Patients and Control Patients' Sydney M . Finegold,2 Dennis J . Flora, Howard R . Attebery, and Vera L. Sutter Medical Service, Wadsworth Hospital Center, Veterans Administration and Department ofMedicine, University ofCalifornia at Los Angeles School of Medicine, Los Angeles, California 9tXl24 Summary and villous adenomata are rare in populations with a low incidence of colon cancer and that they are found with the Feces from 25 subjects with colonic polyps (multiple highest incidence in areas where colon cancer has the adenomatous, large single, or single with atypia) and from highest prevalence. Berge et a!. (3) note a close association 25 matched control subjects were studied by detailed between polyps and carcinoma, both tending to occur in the quantitative aerobic and anaerobic techniques, using a large same distribution. Fifty-nine % of polypoid tumors more battery of culture media and several atmospheric condi than 10 mm in diameter were carcinomas, as were 16.9% of tions. Over 55% of organisms detected on microscopic count those 5 to 10 mm in size. were recovered anaerobically. In several cases, there were There has been much speculation on interrelationships significantly different numbers of organisms of specific between diet, intestinal bacteria, intestinal polyps, and types recovered from the two different populations studied. carcinoma (I, 2, 6—8). However, these differed from organisms with “statistical The present study was designed to compare the fecal significance― noted in a previous study from this laboratory bacterial flora of 25 patients with colonic polyps (chiefly involving two different diet groups (Japanese Americans on multiple adenomatous polyps) with that of 25 subjects either a Japanese or a Western diet). -

Common Commensals
Common Commensals Actinobacterium meyeri Aerococcus urinaeequi Arthrobacter nicotinovorans Actinomyces Aerococcus urinaehominis Arthrobacter nitroguajacolicus Actinomyces bernardiae Aerococcus viridans Arthrobacter oryzae Actinomyces bovis Alpha‐hemolytic Streptococcus, not S pneumoniae Arthrobacter oxydans Actinomyces cardiffensis Arachnia propionica Arthrobacter pascens Actinomyces dentalis Arcanobacterium Arthrobacter polychromogenes Actinomyces dentocariosus Arcanobacterium bernardiae Arthrobacter protophormiae Actinomyces DO8 Arcanobacterium haemolyticum Arthrobacter psychrolactophilus Actinomyces europaeus Arcanobacterium pluranimalium Arthrobacter psychrophenolicus Actinomyces funkei Arcanobacterium pyogenes Arthrobacter ramosus Actinomyces georgiae Arthrobacter Arthrobacter rhombi Actinomyces gerencseriae Arthrobacter agilis Arthrobacter roseus Actinomyces gerenseriae Arthrobacter albus Arthrobacter russicus Actinomyces graevenitzii Arthrobacter arilaitensis Arthrobacter scleromae Actinomyces hongkongensis Arthrobacter astrocyaneus Arthrobacter sulfonivorans Actinomyces israelii Arthrobacter atrocyaneus Arthrobacter sulfureus Actinomyces israelii serotype II Arthrobacter aurescens Arthrobacter uratoxydans Actinomyces meyeri Arthrobacter bergerei Arthrobacter ureafaciens Actinomyces naeslundii Arthrobacter chlorophenolicus Arthrobacter variabilis Actinomyces nasicola Arthrobacter citreus Arthrobacter viscosus Actinomyces neuii Arthrobacter creatinolyticus Arthrobacter woluwensis Actinomyces odontolyticus Arthrobacter crystallopoietes -

C-Di-AMP Signaling Is Required for Bile Salts Resistance and Long-Term
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.23.457418; this version posted August 23, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Title: c-di-AMP signaling is required for bile salts resistance and long-term 2 colonization by Clostridioides difficile 3 Authors: Marine Oberkampf1†, Audrey Hamiot1‡†, Pamela Altamirano-Silva2, Paula Bellés- 4 Sancho1§, Yannick D. N. Tremblay1¶, Nicholas DiBenedetto3, Roland Seifert4, Olga 5 Soutourina5, Lynn Bry3,6, Bruno Dupuy1* and Johann Peltier1,5* 6 7 Affiliations: 8 1. Laboratoire Pathogenèse des Bactéries Anaérobies, CNRS-2001, Institut Pasteur, 9 Université de Paris, F-75015 Paris, France. 10 2. Centro de Investigación en Enfermedades Tropicales, Facultad de Microbiología, 11 Universidad de Costa Rica, San José, Costa Rica 12 3. Massachusetts Host-Microbiome Center, Dept. Pathology, Brigham & Women’s Hospital, 13 Harvard Medical School, Boston, MA. 14 4. Institute of Pharmacology & Research Core Unit Metabolomics, Hannover Medical 15 School, Hannover, Germany. 16 5. Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 17 91198, Gif-sur-Yvette, France 18 6. Clinical Microbiology Laboratory, Department of Pathology, Brigham & Women’s 19 Hospital, Boston, MA. 20 21 * Co-corresponding authors. Emails: [email protected]; [email protected] 22 saclay.fr 23 24 † These authors contributed equally to the studies undertaken. 25 ‡ Present address: UMR UMET, INRA, CNRS, Univ. Lille 1, 59650 Villeneuve d'Ascq, 26 France. -

Optimization and Production of Hyaluronidase by Streptococcus
lar Rese cu ar le ch o & m o T i h B e f r Journal of a Mahesh et al., J Biomol Res Ther 2012, 1:1 o p l e a u n t r i DOI: 10.4172/2167-7956.1000101 c u s o J ISSN: 2167-7956 Bioresearch Communications Research Article Open Access Optimization and Production of Hyaluronidase by Streptococcus mitis MTCC 2695 Narayanan Mahesh1*, Srinivasan Balakumar1, R Parkavi1, Arunadevi Ayyadurai1 and Rangarajan Vivek2 1Department of Chemistry and Biosciences, Srinivasa Ramanujan Centre, SASTRA University, Kumbakonam – 612001, India 2Department of Biotechnology, Indian Institute of Technology, Kharagpur-721 302, India Abstract Hyaluronidase was produced by submerged fermentation from Streptococcus mitis. The possibility of using Streptococcus mitis for enzyme production has been recently investigated. In this study, the physical and nutritional parameters were optimized to improve the production of hyaluronidase by Streptococcus mitis and it was accessed. Maximum production of hyaluronidase was obtained when 5% starch supplemented as carbon source achieved by employing (98.7U/ml) and followed by ammonium chloride (140.4U/ml) incubation period about 48 hours showed (108.9U/ml) and temperature at 37°C showed (179.9U/ml). The maximum enzyme yield on pH 4 is (110.7U/ml). The production of hyaluronidase by means of immobilized Streptococcus mitis was evaluated and a maximum production was obtained with the medium was inoculated with 100 beads (591U/ml) which was more than that of mobilized cells. Keywords: Hyaluronidase; Streptococcus mitis; Submerged fermen- prevents their being recognized by phagocytes of the immune system tation; Hyaluronan; Immobilization [11,28]. -

Identification of Clinically Relevant Streptococcus and Enterococcus
pathogens Article Identification of Clinically Relevant Streptococcus and Enterococcus Species Based on Biochemical Methods and 16S rRNA, sodA, tuf, rpoB, and recA Gene Sequencing Maja Kosecka-Strojek 1,* , Mariola Wolska 1, Dorota Zabicka˙ 2 , Ewa Sadowy 3 and Jacek Mi˛edzobrodzki 1 1 Department of Microbiology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, 30-387 Krakow, Poland; [email protected] (M.W.); [email protected] (J.M.) 2 Department of Molecular Microbiology, National Medicines Institute, 00-725 Warsaw, Poland; [email protected] 3 Department of Epidemiology and Clinical Microbiology, National Medicines Institute, 00-725 Warsaw, Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-12-664-6365 Received: 13 October 2020; Accepted: 9 November 2020; Published: 11 November 2020 Abstract: Streptococci and enterococci are significant opportunistic pathogens in epidemiology and infectious medicine. High genetic and taxonomic similarities and several reclassifications within genera are the most challenging in species identification. The aim of this study was to identify Streptococcus and Enterococcus species using genetic and phenotypic methods and to determine the most discriminatory identification method. Thirty strains recovered from clinical samples representing 15 streptococcal species, five enterococcal species, and four nonstreptococcal species were subjected to bacterial identification by the Vitek® 2 system and Sanger-based sequencing methods targeting the 16S rRNA, sodA, tuf, rpoB, and recA genes. Phenotypic methods allowed the identification of 10 streptococcal strains, five enterococcal strains, and four nonstreptococcal strains (Leuconostoc, Granulicatella, and Globicatella genera). The combination of sequencing methods allowed the identification of 21 streptococcal strains, five enterococcal strains, and four nonstreptococcal strains. -

Title Layout
Antibiotics – an update on recently approved and investigational drugs Jenner Minto, Pharm.D. Disclosures • Nothing to disclose Learning Objectives • Describe the use of new antibacterial agents in clinical practice • Describe the therapeutic potential of antibiotics currently in development Assessment Questions 1. Which of the following statements regarding new antibacterial agents is true? A. Most are FDA approved to treat a broad range of infections B. Recently approved antibiotics are likely to become first-line agents C. Many have a novel mechanism of action D. Most are reserved for infections caused by organisms that are resistant to existing antibiotics 2. The majority of antibiotics currently in development target which pathogen(s)? A. Gram-positive organisms B. Gram-negative ESKAPE pathogens C. Drug-resistant Neisseria gonorrheae D. Drug-resistant Clostridioides difficile Recently Approved Antibiotics 2018 Approvals Plazomicin (Zemdri®) Evracycline (Xerava®) Sarecycline (Seysara®) Omadacycline (Nuzyra®) Rifamycin (Aemcolo®) 2019 Approvals Imipenem, cliastatin, relebactam (Recabrio®) Pretomanid Lefamulin (Xenleta®) Cefiderocol (Fetroja®) Plazomicin (Zemdri®) Approved • June, 2019 Indications • Complicated urinary tract infections (cUTIs), including pyelonephritis, caused by: E. coli, K. pneumoniae, P. mirabilis, Enterobacter cloacae *Reserved for patients ≥ 18 years of age with limited or no alternative treatment options https://zemdri.com/ https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/210303orig1s000lbl.pdf -

Streptococcus Mitis Strains Causing Severe Clinical Disease in Cancer Patients Samuel A
RESEARCH Streptococcus mitis Strains Causing Severe Clinical Disease in Cancer Patients Samuel A. Shelburne, Pranoti Sahasrabhojane, Miguel Saldana, Hui Yao, Xiaoping Su, Nicola Horstmann, Erika Thompson, and Anthony R. Flores The genetically diverse viridans group streptococci in species designation and classification schemes over time (VGS) are increasingly recognized as the cause of a va- (7). From a clinical microbiology laboratory standpoint, riety of human diseases. We used a recently developed automated systems have considerable limitations in VGS multilocus sequence analysis scheme to define the species species identification (8,9). The problematic nature of VGS of 118 unique VGS strains causing bacteremia in patients species assignment also extends to16S rRNA sequencing, with cancer; Streptococcus mitis (68 patients) and S. ora- the most widely used genetic tool for species identification lis (22 patients) were the most frequently identified strains. Compared with patients infected with non–S. mitis strains, in clinical and research settings (9,10). patients infected with S. mitis strains were more likely to Outcomes for patients with VGS bacteremia are highly have moderate or severe clinical disease (e.g., VGS shock variable: some patients have minimal symptoms, and oth- syndrome). Combined with the sequence data, whole-ge- ers have a severe infection characterized by hypotension nome analyses showed that S. mitis strains may more pre- and acute respiratory distress syndrome (11). The severe cisely be considered as >2 species. Furthermore, we found infections have been termed VGS shock syndrome (12). that multiple S. mitis strains induced disease in neutrope- Numerous studies have examined the species distribution nic mice in a dose-dependent fashion. -

HIGHLIGHTS of PRESCRIBING INFORMATION These Highlights Do
HIGHLIGHTS OF PRESCRIBING INFORMATION • Dosage in Pediatric Patients (1 Month of Age to 16 Years): 20 to 40 These highlights do not include all the information needed to use mg/kg/day in 3 or 4 equal doses by intravenous infusion. (2.3) • Alternative Pediatric Patients Dosing: 350 mg/m2/day for serious CLINDAMYCIN IN 0.9% SODIUM CHLORIDE injection safely and 2 effectively. See full prescribing information for CLINDAMYCIN IN infections and 450 mg/m /day for more severe infections. (2.3) 0.9% SODIUM CHLORIDE injection. • Dosage in Neonates (Less than 1 Month of Age): 15 to 20 mg/kg/day in 3 to 4 equal doses by intravenous infusion. (2.3) CLINDAMYCIN IN 0.9% SODIUM CHLORIDE injection, for --------------------- DOSAGE FORMS AND STRENGTHS --------------------- intravenous use Each 50 mL of Clindamycin in 0.9% Sodium Chloride Injection, Initial U.S. Approval: 1989 300 mg/50 mL (6 mg/mL), 600 mg/50 mL (12 mg/mL), and 900 mg/50 mL (18 mg/mL) contains 300 mg, 600 mg, or 900 mg clindamycin, respectively WARNING: CLOSTRIDIOIDES DIFFICILE-ASSOCIATED (as clindamycin phosphate, USP), in a single-dose GALAXY container. (3) DIARRHEA (CDAD) and COLITIS ------------------------------ CONTRAINDICATIONS ----------------------------- See full prescribing information for complete boxed warning. Individuals with a history of hypersensitivity to preparations containing clindamycin or lincomycin. (4) Clostridioides difficile-associated diarrhea (CDAD) has been reported with use of nearly all antibacterial agents, including Clindamycin in ----------------------- WARNINGS AND PRECAUTIONS ----------------------- 0.9% Sodium Chloride Injection and may range in severity from mild • Anaphylactic shock and anaphylactic reactions have been reported. (5.2) diarrhea to fatal colitis. -

Metabolic Changes Associated with Adaptive Resistance to Daptomycin in Streptococcus Mitis-Oralis
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications -- Chemistry Department Published Research - Department of Chemistry 2020 Metabolic changes associated with adaptive resistance to daptomycin in Streptococcus mitis-oralis Allison Parrett Joseph M. Reed Stewart G. Gardner Nagendra N. Mishra Arnold S. Bayer See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/chemfacpub Part of the Analytical Chemistry Commons, Medicinal-Pharmaceutical Chemistry Commons, and the Other Chemistry Commons This Article is brought to you for free and open access by the Published Research - Department of Chemistry at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Faculty Publications -- Chemistry Department by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Allison Parrett, Joseph M. Reed, Stewart G. Gardner, Nagendra N. Mishra, Arnold S. Bayer, Robert Powers, and Greg A. Somerville Parrett et al. BMC Microbiology (2020) 20:162 https://doi.org/10.1186/s12866-020-01849-w RESEARCH ARTICLE Open Access Metabolic changes associated with adaptive resistance to daptomycin in Streptococcus mitis-oralis Allison Parrett1, Joseph M. Reed2,3, Stewart G. Gardner2,4, Nagendra N. Mishra5,6, Arnold S. Bayer5,6, Robert Powers1,7* and Greg A. Somerville2* Abstract Background: Viridans group streptococci of the Streptococcus mitis-oralis subgroup are important endovascular pathogens. They can rapidly develop high-level and durable non-susceptibility to daptomycin both in vitro and in vivo upon exposure to daptomycin. Two consistent genetic adaptations associated with this phenotype (i.e., mutations in cdsA and pgsA) lead to the depletion of the phospholipids, phosphatidylglycerol and cardiolipin, from the bacterial membrane. -

Microbiology and Clinical Characteristics of Viridans Group Streptococci in Patients with Cancer
braz j infect dis 2018;22(4):323–327 The Brazilian Journal of INFECTIOUS DISEASES www.elsevier.com/locate/bjid Original article Microbiology and clinical characteristics of viridans group streptococci in patients with cancer Fuensanta Guerrero-Del-Cueto a, Cyntia Ibanes-Gutiérrez a, Consuelo Velázquez-Acosta b, Patricia Cornejo-Juárez a, Diana Vilar-Compte a,∗ a Instituto Nacional de Cancerología, Departamento de Enfermedades Infecciosas, Ciudade de México, Mexico b Instituto Nacional de Cancerología, Laboratorio de Microbiología, Ciudade de México, Mexico article info abstract Article history: This study assessed the microbiology, clinical syndromes, and outcomes of oncologic Received 22 February 2018 patients with viridans group streptococci isolated from blood cultures between January 1st, Accepted 12 June 2018 2013 and December 31st, 2016 in a referral hospital in Mexico using the Bruker MALDI Bio- Available online 17 July 2018 typer. Antimicrobial sensitivity was determined using BD Phoenix 100 according to CLSI M100 standards. Clinical information was obtained from medical records and descriptive Keywords: analysis was performed. ± Viridans group streptococci Forty-three patients were included, 22 females and 21 males, aged 42 17 years. Twenty Bacteremia (46.5%) patients had hematological cancer and 23 (53.5%) a solid malignancy. The viridans Bloodstream infection group streptococci isolated were Streptococcus mitis, 20 (46.5%); Streptococcus anginosus,14 Cancer (32.6%); Streptococcus sanguinis, 7 (16.3%); and Streptococcus salivarius, 2 (4.7%). The main Mucosal damage risk factors were pyrimidine antagonist chemotherapy in 22 (51.2%) and neutropenia in 19 (44.2%) cases, respectively. Central line associated bloodstream infection was diagnosed in 18 (41.9%) cases. -

Microbiology 2020 MLE-M2
MEDICAL LABORATORY EE VV AA LL UU AA TT II OO NN PARTICIPANT SUMMARY 2 • 0 • 2 • 0 Microbiology 2020 MLE-M2 Total Commitment to Education and Service Provided by ACP, Inc. Table of Contents Evaluation Criteria ................................................... 2 Microbiology MRSA Culture ............................................................ 3 GC (Antigen Detection) .............................................. 22 Strep A Antigen Detection ......................................... 4 Cryptosporidium Antigen Detection ......................... 24 Miscellaneous Cultures ............................................. 8 Giardia lamblia Antigen Detection ............................. 25 Throat Culture ............................................................ 9 RSV Antigen Detection ............................................... 26 Urine Culture ............................................................ 10 Influenza A Antigen Detection ................................... 28 Gram Stain & Morphology ................................... 10 Influenza B Antigen Detection ................................... 30 Antimicrobial Susceptibility Testing ...................... 12 Clostridioides difficile Antigen Detection ................. 32 Genital Culture ......................................................... 13 Rotavirus Antigen Detection ...................................... 33 Gram Stain & Morphology ................................... 13 Legionella Antigen Detection..................................... 34 Colony Count/Urine Presumptive -

Streptococcus Mitis and S. Oralis Lack a Requirement for Cdsa, the Enzyme Required For
AAC Accepted Manuscript Posted Online 21 February 2017 Antimicrob. Agents Chemother. doi:10.1128/AAC.02552-16 Copyright © 2017 Adams et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license. 1 Streptococcus mitis and S. oralis lack a requirement for CdsA, the enzyme required for 2 synthesis of major membrane phospholipids in bacteria 3 Hannah M. Adams1, Luke R. Joyce1, Ziqiang Guan2, Ronda L. Akins1,3, and Kelli L. Palmer1,* 4 Department of Biological Sciences, The University of Texas at Dallas, Richardson, Texas, Downloaded from 5 USA1, Department of Biochemistry, Duke University Medical Center, Durham, NC2, Methodist 6 Charlton Medical Center, Dallas, Texas, USA3 7 8 http://aac.asm.org/ 9 Running title: Daptomycin resistance in S. mitis and S. oralis 10 11 *Corresponding author on February 23, 2017 by DUKE UNIV 12 E-mail: [email protected] (KLP) 13 14 1 15 ABSTRACT 16 Synthesis and integrity of the cytoplasmic membrane is fundamental to cellular life. 17 Experimental evolution studies have hinted at unique physiology in the Gram-positive bacteria 18 Streptococcus mitis and S. oralis. These organisms commonly cause bacteremia and infectious Downloaded from 19 endocarditis (IE) but are rarely investigated in mechanistic studies of physiology and evolution. 20 Unlike other Gram-positive pathogens, high-level (MIC ≥ 256 μg/mL) daptomycin resistance 21 rapidly emerges in S. mitis and S. oralis after a single drug exposure. In this study, we find that 22 inactivating mutations in cdsA are associated with high-level daptomycin resistance in S.