Rub Some Dirt on It: How the Soil Microbiome Can Restore Plants
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Senecio Jacobaea in Northern California, an Enduring Success
ENTOMOPHAGA 35 (I), 1990, 71-77 BIOLOGICAL CONTROL OF SENECIO JACOBAEA IN NORTHERN CALIFORNIA, AN ENDURING SUCCESS R. W. PEMBERTON (t) & C. E. TURNER Rangeland Insect Lab, USDA-ARS, Montana State University, Bozeman, Montana 59717 Biological Control of Weeds Western Regional Research Center USDA-ARS, Albany, California 94710 Seneciojacobaea, a poisonous weed from Eurasia, was brought under successful biological control in the Ft. Bragg, California area by 1976, through the combined action of the defoliating cinnabar moth (Tyriajacobaeae) and a root feeding flea beetle (Longitarsusjacobaeae). In 1987, 4 previously infested Ft. Bragg sites (3 sites where control had been documented and another unstudied site) were examined. Senecio jacobaea densities at these sites were 0.0, 0.0, 0.01 and 0.18 plants/m 2, indicating both continued and improved control of the weed. The flea beetle and the cinnabar moth both persist at the sites, despite very low numbers of S.jacobaea plants. The control of S. jacobaea has resulted in the return of near natural vegetation at the 2 coastal prairie sites and regained productivity at the 2 pasture sites. KEY-WORDS : Biological control, cinnabar moth, Longitarsusjacobaeae, poiso- nous plant, Seneciojacobaea, tansy ragwort, Tyria jacobaeae. INTRODUCTION THE WEED PROBLEM Tansy ragwort, (Senecio jacobaea L. : Asteraceae) is a biennial or short lived perennial herb that is native to Europe eastward to Siberia (Harper & Wood, 1957). The plant is an introduced weed in New Zealand, Australia, South Africa, Argentina, Brazil, Canada, and the United States (Holm et al., 1979). Tansy ragwort contains toxic pyrrolizidine alkaloids which cause liver damage and death to cattle and horses that ingest it (Kingsbury, 1964 ; Harris et aL, 1984). -

Asteraceae, Senecioneae), with Nomenclatural Notes on This Name
Phytotaxa 234 (3): 271–279 ISSN 1179-3155 (print edition) www.mapress.com/phytotaxa/ PHYTOTAXA Copyright © 2015 Magnolia Press Article ISSN 1179-3163 (online edition) http://dx.doi.org/10.11646/phytotaxa.234.3.8 Molecular and cytogenetic confirmation of the hybrid origin of Jacobaea ×mirabilis (Asteraceae, Senecioneae), with nomenclatural notes on this name GEMMA MAS DE XAXARS1, ALAIN FRIDLENDER2, TERESA GARNATJE3 & JOAN VALLÈS1 1 Laboratori de Botànica - Unitat Associada CSIC, Facultat de Farmàcia, Universitat de Barcelona. Avinguda Joan XXIII s/n, 08028 Barcelona (Catalonia, Spain); [email protected], [email protected] 2 Faculté des Sciences, Université de Provence - AMU, case 75, 3 place Victor Hugo, 13331 Marseille Cedex 3 (France); [email protected] 3 Institut Botànic de Barcelona (IBB-CSIC-ICUB), Passeig del Migdia s/n, 08038 Barcelona (Catalonia, Spain); [email protected] Abstract Jacobaea ×mirabilis is a hybrid taxon, formerly included in the genus Senecio, originating from J. adonidifolia and J. leu- cophylla as progenitors. It inhabits a few French mountain locations in the Pyrenees and in the Massif Central. In this paper, we characterize the hybrid and its parental taxa using molecular phylogenetic and cytogenetic (genome size) methods. These data are useful to confirm the hybrid status of the studied taxon. Additionally, we clarify nomenclatural issues connected with this species and fulfil the conditions for valid publication of its name in Jacobaea. The name of this hybrid is also lectotypified. Key words: genome size, homoploid hybridization, nomenclatural combination, phylogeny, Senecio ×mirabilis, typification Introduction Hybridization has classically been and is still recognized as a powerful evolutionary mechanism in plants, particularly in diversification and speciation through genetic exchange (Anderson 1949; Stebbins 1959; Arnold 2006). -
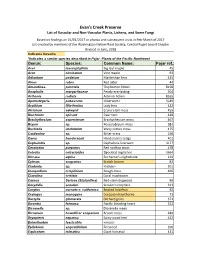
Evans-Creek-Plant-List.Pdf
Evan's Creek Preserve List of Vascular and Non‐Vascular Plants, Lichens, and Some Fungi Based on findings on 01/01/2017 or photos and subsequent visits in Feb‐March of 2017 List created by members of the Washington Native Plant Society, Central Puget Sound Chapter Revised in June, 2018 Indicates invasive *Indicates a similar species described in Pojar -Plants of the Pacific Northwest Genus: Species: Common Name: Pojar ref.: Acer macrophyllum Big leaf maple 45 Acer circinatum Vine maple 93 Adiantum pedatum Maidenhair fern 425 Alnus rubra Red alder 44 Amandinea punctata Tiny button lichen B158 Anaphalis margaritaceae Pearly everlasting 304 Arthonia radiata Asterisk lichen B165 Apometzgeria pubescens <liverwort> SL49 Arythium filix-femina Lady fern 422 Atrichum selwynii Crane's‐bill moss 455 Blechnum spicant Deer fern 420 Brachythecium asperrimum Brachythecium moss 467 Bryum sp. Rossulobryum moss S82 Buckiella undulatum Wavy cotton moss 475 Cardamine sp. Bitter‐cress 146 Carex hendersonii Henderson's sedge 401 Cephalozia sp. Cephalozia liverwort SL77 Ceratodon purpurus Red rooftop moss 478 Cetrelia cetrarioides Speckled rag lichen M64 Circaea alpina Enchanter's nightshade 210 Cytisus scoparius Scotch broom 83 Cladonia sp. <lichen> 501 Claopodium crispifoium Rough moss 466 Clavulina cristata Coral mushroom Cornus Sericea (Stolonifera) Red‐stem dogwood 90 Corydalis scouleri Scouler's corydalis 313 Corylus cornuta v. californica Beaked hazelnut 92 Crategus monogyna European hawthorne 73 Dactylis glomerata Orchard grass 371 Dicentra formosa -

Asteraceae) from Turkey: Senecio Inops Boiss
Turk J Bot 33 (2009) 285-289 © TÜBİTAK Research Article doi:10.3906/bot-0812-3 A new taxon of Senecio (Asteraceae) from Turkey: Senecio inops Boiss. & Balansa subsp. karamanicus Hamzaoğlu & Budak Ergin HAMZAOĞLU1,*, Ümit BUDAK1, Ahmet AKSOY2 1Bozok University, Faculty of Science and Arts, Department of Biology, 66200 Yozgat - TURKEY 2Erciyes University, Faculty of Science and Arts, Department of Biology, 38039 Kayseri - TURKEY Received: 17.12.2008 Accepted: 02.06.2009 Abstract: Senecio inops Boiss. & Balansa subsp. karamanicus Hamzaoğlu & Budak (Asteraceae, Senecioneae) is described as a new subspecies from Karaman Province (Inner/South Anatolia). A Latin diagnosis, a taxonomic description, an illustration of the new subspecies, geographical distribution, and some comments on its affinity withSenecio inops Boiss. & Balansa subsp. inops are given. Key words: Asteraceae, Karaman, Senecio, taxonomy, Turkey Türkiye’den Senecio’nun (Asteraceae) yeni bir taksonu: Senecio inops Boiss. & Balansa subsp. karamanicus Hamzaoğlu & Budak Özet: Senecio inops Boiss. & Balansa subsp. karamanicus Hamzaoğlu & Budak (Asteraceae, Senecioneae) Karaman ilinden yeni bir alttür olarak tanımlandı (İç/Güney Anadolu). Yeni alttürün Latince kısa ayrımı, taksonomik betimlemesi, resmi, coğrafik yayılışı ve Senecio inops Boiss. & Balansa subsp. inops ile yakınlığı hakkında bazı yorumlar verildi. Anahtar sözcükler: Asteraceae, Karaman, Senecio, taksonomi, Türkiye Introduction infrageneric classification of Senecio difficult, and Senecioneae is one of the largest tribes of therefore, the evolutionary history of this genus is still Asteraceae, comprising about 150 genera and 3000 poorly known (Jeffrey et al., 1977; Bremer, 1994; species. Senecio L. is one of about 50 plant genera that Vincent, 1996; Mabberley, 1997). contain over 500 species. The extent of the genus The generic and infrageneric concepts of Senecio (about 1500 species) has made attempts at s.l. -

Frequent Gain and Loss of Pyrrolizidine Alkaloids in the Evolution of Senecio Section Jacobaea (Asteraceae)
PHYTOCHEMISTRY Phytochemistry 66 (2005) 1285–1295 www.elsevier.com/locate/phytochem Frequent gain and loss of pyrrolizidine alkaloids in the evolution of Senecio section Jacobaea (Asteraceae) Pieter B. Pelser a,b,*, Helene de Vos c, Claudine Theuring d, Till Beuerle d, Klaas Vrieling c, Thomas Hartmann d,* a Department of Botany, Miami University, Oxford, OH 45056, USA b Nationaal Herbarium Nederland, Universiteit Leiden Branch, P.O. Box 9514, 2300 RA Leiden, The Netherlands c Institute of Biology, Leiden University, P.O. Box 9516, 2300 RA Leiden, The Netherlands d Institut fu¨r Pharmazeutische Biologie, Technische Universita¨t Braunschweig, Mendelssohnstrasse 1, D-38106 Braunschweig, Germany Received 18 November 2004; received in revised form 12 January 2005 Available online 17 May 2005 Abstract Pyrrolizidine alkaloids (PAs) of the macrocyclic senecionine type are secondary metabolites characteristic for most species of the genus Senecio (Asteraceae). These PAs are deterrent and toxic to most vertebrates and insects and provide plants with a chemical defense against herbivores. We studied the PA composition of 24 out of 26 species of Senecio section Jacobaea using GC-MS. The PA profiles of eight of these species have not been studied before and additional PAs were identified for most other species that were included in previous studies. With one exception (senecivernine) all 26 PAs identified in sect. Jacobaea can be regarded as derivatives of the biosynthetic backbone structure senecionine. Based on the PA profiles of the species of sect. Jacobaea and the results of pre- vious tracer studies, we constructed two hypothetical biosynthetic scenarios of senecionine diversification. Both scenarios contain two major reactions: the conversion of the necine base moiety retronecine into the otonecine moiety and site-specific epoxidations within the necic acid moiety. -

Guidebook to Invasive Nonnative Plants of the Elwha Watershed Restoration
Guidebook to Invasive Nonnative Plants of the Elwha Watershed Restoration Olympic National Park, Washington Cynthia Lee Riskin A project submitted in partial fulfillment of the requirements for the degree of Master of Environmental Horticulture University of Washington 2013 Committee: Linda Chalker-Scott Kern Ewing Sarah Reichard Joshua Chenoweth Program Authorized to Offer Degree: School of Environmental and Forest Sciences Guidebook to Invasive Nonnative Plants of the Elwha Watershed Restoration Olympic National Park, Washington Cynthia Lee Riskin Master of Environmental Horticulture candidate School of Environmental and Forest Sciences University of Washington, Seattle September 3, 2013 Contents Figures ................................................................................................................................................................. ii Tables ................................................................................................................................................................. vi Acknowledgements ....................................................................................................................................... vii Introduction ....................................................................................................................................................... 1 Bromus tectorum L. (BROTEC) ..................................................................................................................... 19 Cirsium arvense (L.) Scop. (CIRARV) -

Woodland Ragwort Senecio Sylvaticus L
woodland ragwort Senecio sylvaticus L. Synonyms: none Other common names: heath groundsel, woodland groundsel Family: Asteraceae Invasiveness Rank: 41 The invasiveness rank is calculated based on a species’ ecological impacts, biological attributes, distribution, and response to control measures. The ranks are scaled from 0 to 100, with 0 representing a plant that poses no threat to native ecosystems and 100 representing a plant that poses a major threat to native ecosystems. Description Similar species: Hultén documented 11 native Senecio Woodland ragwort is a taprooted annual plant that species in Alaska. Four other Senecio species are grows up to 80 cm tall. The plant is covered in tracked non-native plants in Alaska. The non-native abundant, fine hairs. Stems are single and unbranched or common groundsel (Senecio vulgaris) can be confused branched. Leaves are obovate to oblong, 3 to 7 cm long, with woodland ragwort. However, woodland ragwort 1 to 3 cm wide, one or two times pinnate, and alternate. can be distinguished by the complete absence or Leaves are petiolated with irregular teeth and tapered presence of a few, green-tipped, outer reduced bases; upper leaves are clasping. Each stem produces a involucral bracts at the base of the flower head. In cluster of 12 to 24 flower heads. Flower heads have 0 to contrast, common groundsel has reduced outer 15 ray florets, which, when present are each 1 to 2 mm involucral bracts at the base of the flower head long. Involucral bracts are narrow and 5 to 7 mm long characterized by conspicuous black tips. Unlike with short hairs and sometimes with black tips. -

Strong Incongruence Between the ITS Phylogeny and Generic Delimitation in the Nemosenecio-Sinosenecio- Tephroseris Assemblage (Asteraceae: Senecioneae)
Botanical Studies (2009) 50: 435-442. PHYLOGENETICS Strong incongruence between the ITS phylogeny and generic delimitation in the Nemosenecio-Sinosenecio- Tephroseris assemblage (Asteraceae: Senecioneae) Liu-YangWANG1,2,5,PieterB.PELSER3,BertilNORDENSTAM4, and Jian-Quan LIU2,* 1Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Plateau Institute of Biology, Chinese Academy of Sciences, Xining 810001, P.R. China 2Key Laboratory of Arid and Grassland Ecology, School of Life Science, Lanzhou University, Lanzhou 730000, P.R. China 3Oklahoma State University, Botany Department, 104 Life Sciences East Stillwater, Oklahoma 74078-3013, USA 4Swedish Museum of Natural History, P. O. Box 50007, SE-104 05 Stockholm, Sweden 5Graduate University of Chinese Academy of Sciences, Beijing 100049, P.R. China (Received January 9, 2009; Accepted March 9, 2009) ABSTRACT. ThethreegeneraSinosenecio,NemosenecioandTephroserisformacloselyknitgroupnested inthesubtribeTussilagininaeofthetribeSenecioneae(Asteraceae).Thegenericlimitsinthisassemblage remain unclear and need revision. In this study, we analysed sequences of the internal transcribed spacer (ITS) region of nuclear ribosomal DNA available from GenBank and sequenced 19 accessions of an additional 13 species encompassing all three genera. Phylogenetic analyses based on the ITS variation of 27 species in this assemblage and seven species from related genera of the Tussilagininae suggested that neither Sinosenecio norTephroserisismonophyletic.ThesampledspeciesofSinoseneciowerescatteredindifferentcladesor -

Soil Preferences and Morphological Diversity of Goldenrods (Solidago L.) from South-Western Poland
Acta Societatis Botanicorum Poloniae Journal homepage: pbsociety.org.pl/journals/index.php/asbp ORIGINAL RESEARCH PAPER Received: 2013.03.13 Accepted: 2012.11.29 Published electronically: 2013.04.27 Acta Soc Bot Pol 82(2):107–115 DOI: 10.5586/asbp.2013.005 Soil preferences and morphological diversity of goldenrods (Solidago L.) from south-western Poland Magdalena Szymura1*, Tomasz H. Szymura2 1 Department of Agroecosystems and Green Areas Management, Wrocław University of Environmental and Life Sciences, pl. Grunwaldzki 24a, 53-363 Wrocław, Poland 2 Department of Ecology, Biogeochemistry and Environmental Protection, University of Wrocław, Kanonia 6/8, 50-328 Wrocław, Poland Abstract Invasive plants in their new range can differ from their ancestors, including traits ultimately influencing habitat preferences, competitiveness and dispersal ability. In Europe Solidago species are considered as one of the worst invaders of American origin. In this study the frequency of occurrence of Solidago species, their soil preferences and morphological diversity, in Silesia (south- western Poland, Central Europe) were surveyed. On the basis of phytosociological relevés, made using the Braun-Blanquet method, in 75 plots, we determined the composition of species co-occurring with particular Solidago species. The height of ramets, as well as length and width of inflorescences ofSolidago species were measured. We also determined the basic soil properties and noted the presence of trees overshading the ground vegetation. The compositional variation of vegetation and its relation to environmental traits: soil properties (texture, pH, percentage of organic matter, total nitrogen, nitrate, phosphorus, potassium and calcium content) and presence of canopy were analyzed by multivariate ordination methods (CA and CCA). -

Element Stewardship Abstract for Senecio Jacobaea
ELEMENT STEWARDSHIP ABSTRACT for Senecio jacobaea Tansy Ragwort, Tansy Butterweed To the User: Element Stewardship Abstracts (ESAs) are prepared to provide The Nature Conservancy's Stewardship staff and other land managers with current management-related information on those species and communities that are most important to protect, or most important to control. The abstracts organize and summarize data from numerous sources including literature and researchers and managers actively working with the species or community. We hope, by providing this abstract free of charge, to encourage users to contribute their information to the abstract. This sharing of information will benefit all land managers by ensuring the availability of an abstract that contains up-to-date information on management techniques and knowledgeable contacts. Contributors of information will be acknowledged within the abstract and receive updated editions. To contribute information, contact the editor whose address is listed at the end of the document. For ease of update and retrievability, the abstracts are stored on computer at the national office of The Nature Conservancy. This abstract is a compilation of available information and is not an endorsement of particular practices or products. Please do not remove this cover statement from the attached abstract. Authors of this Abstract: Cathy Macdonald, Mary J. Russo (Revision) © THE NATURE CONSERVANCY 1815 North Lynn Street, Arlington, Virginia 22209 (703) 841 5300 The Nature Conservancy Element Stewardship Abstract For Senecio jacobaea I. IDENTIFIERS Common Name: Tansy Ragwort, Tansy Butterweed General Description: The following description of Senecio jacobaea is adapted from Munz and Keck (1973). Senecio jacobaea is a member of the Groundsel Tribe (Senecioneae) of the Sunflower Family (Asteraceae=Compositae). -

Southern Garden History Plant Lists
Southern Plant Lists Southern Garden History Society A Joint Project With The Colonial Williamsburg Foundation September 2000 1 INTRODUCTION Plants are the major component of any garden, and it is paramount to understanding the history of gardens and gardening to know the history of plants. For those interested in the garden history of the American south, the provenance of plants in our gardens is a continuing challenge. A number of years ago the Southern Garden History Society set out to create a ‘southern plant list’ featuring the dates of introduction of plants into horticulture in the South. This proved to be a daunting task, as the date of introduction of a plant into gardens along the eastern seaboard of the Middle Atlantic States was different than the date of introduction along the Gulf Coast, or the Southern Highlands. To complicate maters, a plant native to the Mississippi River valley might be brought in to a New Orleans gardens many years before it found its way into a Virginia garden. A more logical project seemed to be to assemble a broad array plant lists, with lists from each geographic region and across the spectrum of time. The project’s purpose is to bring together in one place a base of information, a data base, if you will, that will allow those interested in old gardens to determine the plants available and popular in the different regions at certain times. This manual is the fruition of a joint undertaking between the Southern Garden History Society and the Colonial Williamsburg Foundation. In choosing lists to be included, I have been rather ruthless in expecting that the lists be specific to a place and a time. -

WO 2016/092376 Al 16 June 2016 (16.06.2016) W P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/092376 Al 16 June 2016 (16.06.2016) W P O P C T (51) International Patent Classification: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, A61K 36/18 (2006.01) A61K 31/465 (2006.01) KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, A23L 33/105 (2016.01) A61K 36/81 (2006.01) MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, A61K 31/05 (2006.01) BO 11/02 (2006.01) PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, A61K 31/352 (2006.01) SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (21) International Application Number: PCT/IB20 15/002491 (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (22) International Filing Date: GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, 14 December 2015 (14. 12.2015) TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, (25) Filing Language: English TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, (26) Publication Language: English LV, MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, (30) Priority Data: SM, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, 62/09 1,452 12 December 201 4 ( 12.12.20 14) US GW, KM, ML, MR, NE, SN, TD, TG).