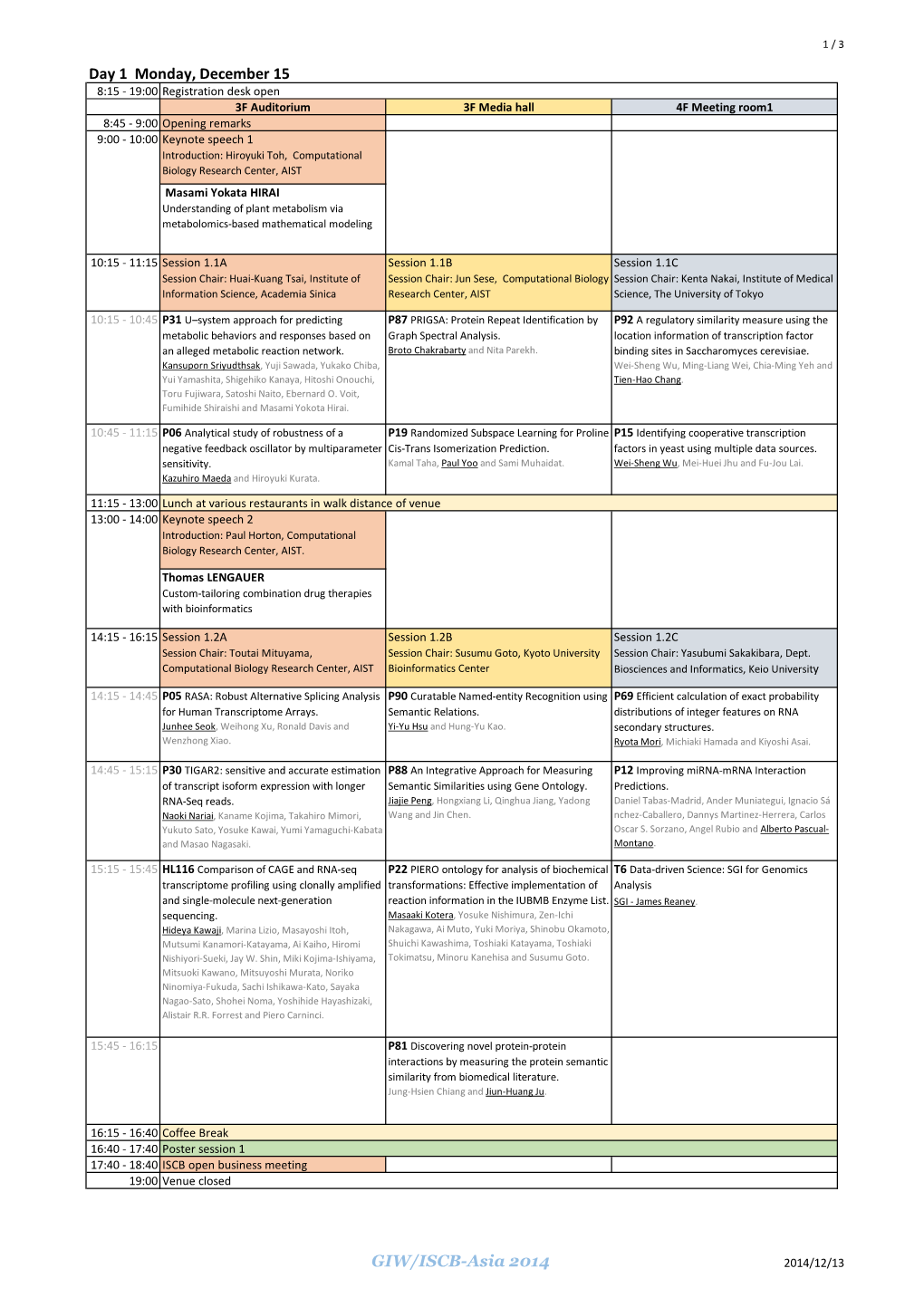
Day 1 Monday, December 15 GIW/ISCB-Asia 2014
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ismb/Eccb 2015
Research Collection Journal Article ISMB/ECCB 2015 Author(s): Moreau, Yves; Beerenwinkel, Niko Publication Date: 2015 Permanent Link: https://doi.org/10.3929/ethz-b-000102416 Originally published in: Bioinformatics 31(12), http://doi.org/10.1093/bioinformatics/btv303 Rights / License: Creative Commons Attribution-NonCommercial 4.0 International This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library Bioinformatics, 31, 2015, i1–i2 doi: 10.1093/bioinformatics/btv303 ISMB/ECCB 2015 Editorial ISMB/ECCB 2015 This special issue of Bioinformatics serves as the proceedings of the 175 external reviewers recruited as sub-reviewers by program com- joint 23rd annual meeting of Intelligent Systems for Molecular mittee members. Table 1 provides a summary of the areas, area Biology (ISMB) and 14th European Conference on Computational chairs and a review summary by area. The conference used a two- Biology (ECCB), which took place in Dublin, Ireland, July 10–14, tier review system—a continuation and refinement of a process that 2015 (http://www.iscb.org/ismbeccb2015). ISMB/ECCB 2015, the begun with ISMB/ECCB 2013 in an effort to better ensure thorough official conference of the International Society for Computational and fair reviewing. Under the revised process, each of the 241 sub- Biology (ISCB, http://www.iscb.org/), was accompanied by nine missions was first reviewed by at least three expert referees, with a Special Interest Group meetings of 1 or 2 days each, and two satel- subset receiving between four and six reviews, as needed. lite meetings. -

Computational Biology and Bioinformatics
Vol. 30 ISMB 2014, pages i1–i2 BIOINFORMATICS EDITORIAL doi:10.1093/bioinformatics/btu304 Editorial This special issue of Bioinformatics serves as the proceedings of The conference used a two-tier review system, a continuation the 22nd annual meeting of Intelligent Systems for Molecular and refinement of a process begun with ISMB 2013 in an effort Biology (ISMB), which took place in Boston, MA, July 11–15, to better ensure thorough and fair reviewing. Under the revised 2014 (http://www.iscb.org/ismbeccb2014). The official confer- process, each of the 191 submissions was first reviewed by at least ence of the International Society for Computational Biology three expert referees, with a subset receiving between four and (http://www.iscb.org/), ISMB, was accompanied by 12 Special eight reviews, as needed. These formal reviews were frequently Interest Group meetings of one or two days each, two satellite supplemented by online discussion among reviewers and Area meetings, a High School Teachers Workshop and two half-day Chairs to resolve points of dispute and reach a consensus on tutorials. Since its inception, ISMB has grown to be the largest each paper. Among the 191 submissions, 29 were conditionally international conference in computational biology and bioinfor- accepted for publication directly from the first round review Downloaded from matics. It is expected to be the premiere forum in the field for based on an assessment of the reviewers that the paper was presenting new research results, disseminating methods and tech- clearly above par for the conference. A subset of 16 papers niques and facilitating discussions among leading researchers, were viewed as potentially in the top tier but raised significant practitioners and students in the field. -

ISMB/ECCB Rebooted: 2015 Brings Major Update to the Meeting Program
MESSAGE FROM ISCB ISMB/ECCB Rebooted: 2015 Brings Major Update to the Meeting Program Christiana N. Fogg1, Diane E. Kovats2* 1 Freelance Science Writer, Kensington, Maryland, United States of America, 2 International Society for Computational Biology, La Jolla, California, United States of America * [email protected] The 23rd Annual International Conference on Intelligent Systems for Molecular Biology and the 14th European Conference on Computational Biology (ISMB/ECCB 2015) is shaping up to be a phenomenal meeting, with world-renowned keynote speakers and changes in the meeting format that aim to make it a more streamlined and user-friendly conference. The joint ISMB/ ECCB conference is held biennially and is the flagship meeting of the International Society for Computational Biology (ISCB). The meeting will take place at the Convention Center Dublin, Ireland, from July 10–14, 2015, and will bring together scientists from across the globe working in a broad range of computational biology—related disciplines, including genomics, structural biology, proteomics, data mining, machine learning, and systems biology. Online registration is open until June 26: https://www.iscb.org/ismbeccb2015-registration. In the past, presentations at the meeting have been organized according to tracks, namely, the Proceedings, Highlights, and Late Breaking Research tracks. In 2015, all oral presentations will be presented in broad thematic areas. As in the past, submissions accepted into the highly selective Proceedings track will be published in a special ISMB/ECCB Proceedings issue of Bioinformatics. Conference co-chairs Alex Bateman, Janet Kelso, and Des Higgins and the conference com- mittee undertook the reorganization effort and came up with five thematic areas: Genes, Pro- teins, Systems, Disease, and Data. -

Curriculum Vitae
Curriculum vitae NAME Martin Kircher BORN 1983 in Erfurt, Germany STUDIES 09/2007 - 07/2011: Dr. rer. nat. (PhD) in Computer Sciences (summa cum laude) at Universität Leipzig, Germany Thesis: Understanding and improving high-throughput sequencing data production and analysis 04/2006 - 07/2007: Master of Science (Honor's Degree) at Saarland University, Germany Computational Molecular Biology Thesis: In silico analysis of microRNA genes on human chr. 14 and mouse chr. 12 10/2003 - 03/2006: Bachelor of Science at Saarland University, Germany Bioinformatics AFFILIATIONS since 04/2012: Jay Shendure, PhD, MD Department of Genome Sciences, University of Washington, Seattle, WA, USA Senior fellow 06/2012 - 11/2015: Center for Mendelian Genomics at the University of Washington, Seattle, WA, USA Lead data analyst 09/2007 - 03/2012: Janet Kelso, PhD & Prof. Dr. Svante Pääbo Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany PhD student (Bioinformatics, Evolutionary Genetics) (09/2007 - 02/2011) Research scientist (Bioinformatics, Evolutionary Genetics) (03/2011 – 03/2012) 05/2007 - 07/2007: Professor Dr. Thomas Lengauer, PhD Max Planck Institute for Informatics, Saarbrücken, Germany Student assistant 09/2006 - 10/2006: Professor Dr. Andreas Zeller Software Engineering Chair, Saarland University, Germany Tutor of practical training in software design 08/2005 - 01/2006: Professor Dr. Hans-Peter Lenhof Center for Bioinformatics, Saarland University, Germany Student assistant VISITS 09/2010 - 11/2010, 11/2009: Dr. Philipp Khaitovich CAS-MPG Partner institute for Computational Biology, Shanghai, China Analysis of Illumina Digital Gene Expression data Consultant for high-throughput sequencing 03/2008: Dr. Dirk Schübeler Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland Measuring DNA methylation using MeDIP [email protected] 1/12 Dr. -

CV Burkhard Rost
Burkhard Rost CV BURKHARD ROST TUM Informatics/Bioinformatics i12 Boltzmannstrasse 3 (Rm 01.09.052) 85748 Garching/München, Germany & Dept. Biochemistry & Molecular Biophysics Columbia University New York, USA Email [email protected] Tel +49-89-289-17-811 Photo: © Eckert & Heddergott, TUM Web www.rostlab.org Fax +49-89-289-19-414 Document: CV Burkhard Rost TU München Affiliation: Columbia University TOC: • Tabulated curriculum vitae • Grants • List of publications Highlights by numbers: • 186 invited talks in 29 countries (incl. TedX) • 250 publications (187 peer-review, 168 first/last author) • Google Scholar 2016/01: 30,502 citations, h-index=80, i10=179 • PredictProtein 1st Internet server in mol. biol. (since 1992) • 8 years ISCB President (International Society for Computational Biology) • 143 trained (29% female, 50% foreigners from 32 nations on 6 continents) Brief narrative: Burkhard Rost obtained his doctoral degree (Dr. rer. nat.) from the Univ. of Heidelberg (Germany) in the field of theoretical physics. He began his research working on the thermo-dynamical properties of spin glasses and brain-like artificial neural networks. A short project on peace/arms control research sketched a simple, non-intrusive sensor networks to monitor aircraft (1988-1990). He entered the field of molecular biology at the European Molecular Biology Laboratory (EMBL, Heidelberg, Germany, 1990-1995), spent a year at the European Bioinformatics Institute (EBI, Hinxton, Cambridgshire, England, 1995), returned to the EMBL (1996-1998), joined the company LION Biosciences for a brief interim (1998), became faculty in the Medical School of Columbia University in 1998, and joined the TUM Munich to become an Alexander von Humboldt professor in 2009. -

Sriram Sankararaman 1/11
Sriram Sankararaman 1/11 SRIRAM SANKARARAMAN 296B Engineering VI Phone: (510) 813-9888 UCLA Email: [email protected] Los Angeles, CA - 90095 www.cs.ucla.edu/~sriram EDUCATION Course in Advanced Bacterial Genetics June 2010 Cold Spring Harbor Laboratory Ph.D. in Computer Science Aug 2004 { May 2010 with a Designated Emphasis in Computational and Genomic Biology University of California, Berkeley B. Tech. in Computer Science and Engineering June 2000 { June 2004 Indian Institute of Technology, Madras POSITIONS AND EMPLOYMENT Associate Professor July 2021 { Assistant Professor Nov 2015 { June 2021 Department of Computer Science, Department of Human Genetics, Department of Computational Medicine, University of California, Los Angeles Post-doctoral fellow Sept 2010 { Nov 2015 Department of Genetics, Harvard Medical School Mentor: David Reich Fellow January { May 2014 Program in Evolutionary Biology and Theory of Computing Simons Institute for the Theory of Computing, Berkeley Intern May{Aug 2006 International Computer Science Institute, Berkeley Mentor: Eran Halperin Intern May{Aug 2005 International Computer Science Institute, Berkeley Mentor: Scott Shenker RESEARCH INTERESTS Computational Biology, Computational/Statistical genomics, Statistical Machine Learning (Probabilistic Graphical Models, Bayesian Statistics). My interests lie at the interface of computer science, statistics and biology. I am interested in developing statistical and computational tools to understand evolution as well as the genetic basis of complex phenotypes. I am also interested in the statistical and inferential challenges posed by the scale of genomic data. DISSERTATION Title: \Statistical Models for Analyzing Human Genetic Variation" Advisors: Prof. Michael Jordan and Prof. Kimmen Sj¨olander Sriram Sankararaman 2/11 ACADEMIC HONORS AND FELLOWSHIPS • NSF Career Award 2020 • Microsoft Investigator Fellowship 2019 • Northrop Grumman Excellence in Teaching Award 2019 • Okawa Foundation Research Grant 2017 • UCLA Hellman Fellow, 2017-2018 • Alfred P. -

Friday (August 4) Intelligent Systems for Molecular Biology
and 2nd Annual AB3C Conference: X-Meeting fortaleza, brazil - auGust 6-10, 2006 friday (auGust 4) Intelligent Systems for Molecular Biology Room A Room B1 Room E1 (2nd Floor) 7:00 a.m. Shuttles depart Hotels to Convention Center 7:30 a.m. - 6:00 p.m. Registration - Hall G 3Dsig Alternative Splicing BOSC 9:00 a.m. - 10:30 a.m. Satellite Meeting SIG SIG 10:30 a.m. - 11:00 a.m. Coffee Break - Satellite & SIG meetings (available outside meeting rooms) 3Dsig Alternative Splicing BOSC 11:00 a.m - 12:00 p.m. Satellite Meeting SIG SIG 12:00 p.m. - 1:00 p.m. Lunch - Satellite and SIG meetings (Hall F) 3Dsig Alternative Splicing BOSC 1:00 p.m. - 3:30 p.m. Satellite Meeting SIG SIG 3:30 p.m. - 4:00 p.m. Coffee Break - Satellite & SIG meetings (available outside meeting rooms) 3Dsig Alternative Splicing BOSC 4:00 p.m. - 6:30 p.m. Satellite Meeting SIG SIG 6:30 p.m. Shuttles depart Convention Center to Hotels 1 and 2nd Annual AB3C Conference: X-Meeting fortaleza, brazil - auGust 6-10, 2006 saturday (auGust 5) Intelligent Systems for Molecular Biology Room A Room B1 Room B2 Room E1 (2nd Floor) 7:00 a.m. Shuttles depart Hotels to Convention Center 7:30 a.m. - 6:00 p.m. Registration - Hall G Joint BioLINK & 3Dsig Alternative Splicing Bio-ontologies BOSC 9:00 a.m. - 10:30 a.m. Satellite Meeting SIG SIG SIG 10:30 a.m. - 11:00 a.m. Coffee Break - Satellite & SIG meetings (available outside meeting rooms) Joint BioLINK & 3Dsig Alternative Splicing Bio-ontologies BOSC 11:00 a.m - 12:00 p.m. -

Full Program Book
1 Welcome On behalf of the ECCB’14 Organizing and Steering Committees we are very happy to welcome you to Strasbourg, Heart of Europe. We hope that you will enjoy the many facets of the conference: keynote lectures, oral communications, workshops and tutorials, demos, posters, booths. This dense and attractive program is intended to be the substrate of fruitful discussions and networking among all of you. We are proud to welcome seven distinguished Keynote Speakers: Nobel Prize laureate Jean-Marie Lehn (Strasbourg University, Nobel Prize in Chemistry in 1987), Patrick Aloy (Institute for Research in Biomedicine, Barcelona, Spain), Alice McHardy (Heinrich Heine University, Düsseldorf and Helmholtz Center for Infection Biology, Braunschweig, Germany), Nada Lavrač (Jožef Stefan Institute and University of Nova Gorica, Slovenia), Ewan Birney (European Bioinformatics Institute, Hinxton, United Kingdom), Doron Lancet (The Weizmann Institute of Science, Rehovot, Israel) and Eric Westhof (Strasbourg University, France). A large variety of Workshops, Tutorials and Satellite Meetings will take place on Saturday, September 6 and Sunday, September 7 to kick off the conference! We would like to stress the interest in the conference by the Council of Europe, whose headquarters are in Strasbourg. An invited talk by Laurence Lwoff, Head of the Bioethics Unit, is scheduled during the opening ceremony, in which she will share the Council of Europe’s concerns about the bio-ethical challenges raised by the usage of biobanks and biomedical data in research and its applications. The Council of Europe will also host the Gala Evening on Tuesday in its nice reception hall and gardens along the Ill river. -
Computational Biology: Moving Into the Future One Click at a Time
MESSAGE FROM ISCB Computational Biology: Moving into the Future One Click at a Time Christiana N. Fogg1*, Diane E. Kovats2 1 Freelance Science Writer, Kensington, Maryland, United States of America, 2 Executive Director, International Society for Computational Biology, La Jolla, California, United States of America * [email protected] Overview Computational biology has grown and matured into a discipline at the heart of biological research. In honor of the tenth anniversary of PLOS Computational Biology, Phil Bourne, Win Hide, Janet Kelso, Scott Markel, Ruth Nussinov, and Janet Thornton shared their memories of the heady beginnings of computational biology and their thoughts on the field’s promising and provocative future. Philip E. Bourne Philip Bourne (Fig 1) began his scientific career in the wet lab, like many of his computational biology contemporaries. He earned his PhD in physical chemistry from the Flinders University of South Australia and pursued postdoctoral training at the University of Sheffield, where he began studying protein structure. Bourne accepted his first academic position in 1995 in the Department of Pharmacology at the University of California, San Diego (UCSD), rose to the rank of professor, and was associate vice chancellor for Innovation and Industry Alliances of the Office of Research Affairs. During his time at UCSD, he built a broad research program that used bioinformatics and systems biology to examine protein structure and function, evolu- tion, drug discovery, disease, and immunology. Bourne also developed the Research Collabora- tory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) and Immune Epitope OPEN ACCESS Database (IEDB), which have become valuable data resources for the research community. -
The University of Chicago Interrogating the 3D Structure of Primate Genomes a Dissertation Submitted to the Faculty of the Divis
THE UNIVERSITY OF CHICAGO INTERROGATING THE 3D STRUCTURE OF PRIMATE GENOMES A DISSERTATION SUBMITTED TO THE FACULTY OF THE DIVISION OF THE BIOLOGICAL SCIENCES AND THE PRITZKER SCHOOL OF MEDICINE IN CANDIDACY FOR THE DEGREE OF DOCTOR OF PHILOSOPHY DEPARTMENT OF HUMAN GENETICS BY ITTAI ETHAN ERES CHICAGO, ILLINOIS DECEMBER 2020 Copyright © 2020 by Ittai Ethan Eres All Rights Reserved Freely available under a CC-BY 4.0 International license \If I am not for myself, who will be for me? But if I am only for myself, who am I? And if not now, when?" Rabbi Hillel Table of Contents LIST OF FIGURES . vi LIST OF TABLES . vii ACKNOWLEDGMENTS . viii ABSTRACT . xi 1 INTRODUCTION . 1 1.1 The evolution of gene regulation . 1 1.2 Gene regulatory evolution insights from comparative primate genomics . 3 1.3 The growing importance of the 3D genome . 11 2 REORGANIZATION OF 3D GENOME STRUCTURE MAY CONTRIBUTE TO GENE REGULATORY EVOLUTION IN PRIMATES . 15 2.1 Abstract . 15 2.2 Introduction . 16 2.3 Results . 18 2.3.1 Inter-species differences in 3D genomic interactions . 19 2.3.2 The relationship between inter-species differences in contacts and gene expression . 26 2.3.3 The chromatin and epigenetic context of inter-species differences in 3D genome structure . 30 2.4 Discussion . 33 2.4.1 Contribution of variation in 3D genome structure to expression diver- gence . 36 2.4.2 Functional annotations . 37 2.5 Materials and methods . 38 2.5.1 Ethics statement . 38 2.5.2 Induced pluripotent stem cells (iPSCs) . -

Bioinformatics
ADVERT ISING & CORPORAT E SERVICES BI OI N FOR M ATI CS | Bioinformatics The leading journal in its field, Bioinformatics publishes the highest quality scientific papers and review articles of interest to academic and industrial researchers. Its main focus is on new developments in genome bioinformatics and computational biology. Two distinct sections within the journal - Discovery Notes and Application Notes - focus on shorter papers, the former reporting biologically interesting discoveries using computational methods, the latter exploring the applications used for experiments. Academic and industrial researchers with an interest in genome bioinformatics and computational biology 24 5.481 International Society for Computational Biology About Oxford University Press's high-quality, peer-reviewed content offers advertisers and sponsors various opportunities to partner with highly credible brands. At Oxford, we have an experienced Corporate Sales global Advertising and Corporate sales force dedicated to serving the needs of our corporate clients and customers. ADVERT ISING & CORPORAT E SERVICES BI OI N FOR M ATI CS | http://academic.oup.com/bioinformatics Overview 3/59 Science: MATHEMATICAL & COMPUTATIONAL BIOLOGY 6/79 Science: BIOCHEMICAL RESEARCH METHODS 17/160 Science: BIOTECHNOLOGY & APPLIED MICROBIOLOGY 2017 Journal Citation Reports® (Clarivate Analytics, 2018) 648743 235321 Janet Kelso Alfonso Valencia Sales Contact Contact your sales manager today to discuss the opportunities available to you. Advertising Sales Manager Reprint Sales Manager Head of Custom Content +44 (0) 7880 008129 +44 (0) 7843 516819 +44 (0) 7702 524839 [email protected] [email protected] [email protected] ADVERT ISING & CORPORAT E SERVICES BI OI N FOR M ATI CS | http://academic.oup.com/bioinformatics Audience Work with us to maximize your exposure to the best in the field. -

Ismb 2010 Organization
Vol. 26 ISMB 2010, pages i2–i6 BIOINFORMATICS doi:10.1093/bioinformatics/btq246 ISMB 2010 ORGANIZATION PROCEEDINGS COMMITTEE CHAIRS Gene Regulation and Transcriptomics Mona Singh, Proceedings Chair, Princeton University, USA Hanah Margalit, The Hebrew University of Jerusalem, Israel Joel S. Bader, Proceedings Co-Chair, Johns Hopkins University, Eric Xing, Carnegie Mellon University, Pittsburgh, USA Baltimore, USA Population Genomics Eleazar Eskin, University of California, Los Angeles, USA Eran Halperin, Tel-Aviv University, Israel PROCEEDINGS AREA CHAIRS Protein Interactions and Molecular Networks Bioimaging Alfonso Valencia, Spanish National Cancer Research Centre, Gene Myers, Howard Hughes Medical Institute, Ashburn, Madrid, Spain USA Roded Sharan, Tel-Aviv University, Israel Robert F. Murphy, Carnegie Mellon University, Pittsburgh, Protein Structure and Function USA Bonnie Berger, Massachusetts Institute of Technology, Cambridge, Databases and Ontologies USA Alex Bateman, Wellcome Trust Sanger Institute, Hinxton, UK Nir Ben-Tal, Tel-Aviv University, Israel Suzanna Lewis, Lawrence Berkeley National Labs, Berkeley, Sequence Analysis USA Michael Brudno, University of Toronto, Canada Disease Models and Epidemiology Cenk Sahinalp, Simon Fraser University, Vancouver, Canada Thomas Lengauer, Max-Planck Institute for Informatics, Text Mining Saarbruken, Germany Andrey Rzhetsky, University of Chicago, USA Yves Moreau, Catholic University of Leuven, Belgium Hagit Shatkay, Queen’s University, Kingston, Canada Evolution and Comparative Genomics