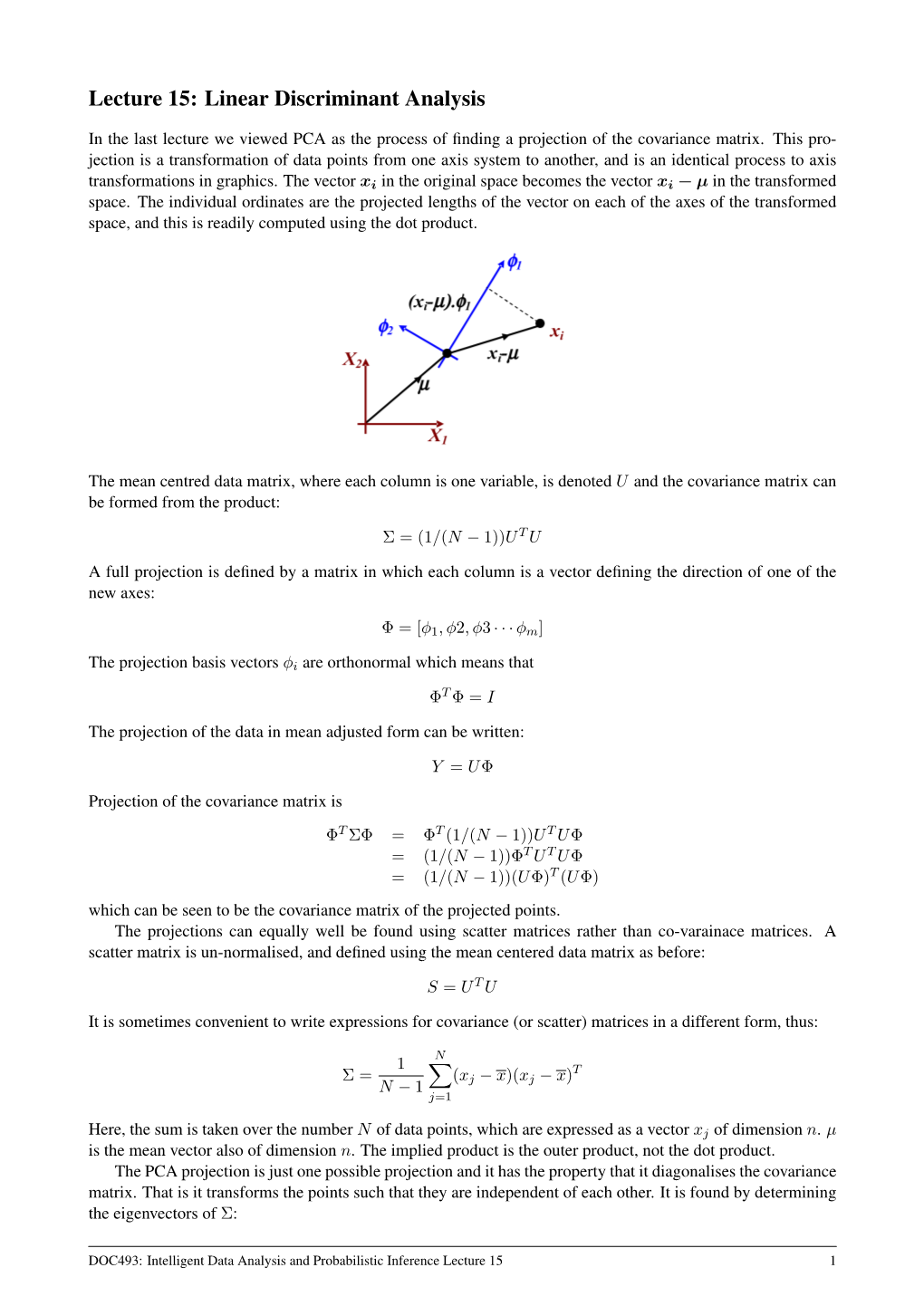
Lecture 15: Linear Discriminant Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Graph Equivalence Classes for Spectral Projector-Based Graph Fourier Transforms Joya A
1 Graph Equivalence Classes for Spectral Projector-Based Graph Fourier Transforms Joya A. Deri, Member, IEEE, and José M. F. Moura, Fellow, IEEE Abstract—We define and discuss the utility of two equiv- Consider a graph G = G(A) with adjacency matrix alence graph classes over which a spectral projector-based A 2 CN×N with k ≤ N distinct eigenvalues and Jordan graph Fourier transform is equivalent: isomorphic equiv- decomposition A = VJV −1. The associated Jordan alence classes and Jordan equivalence classes. Isomorphic equivalence classes show that the transform is equivalent subspaces of A are Jij, i = 1; : : : k, j = 1; : : : ; gi, up to a permutation on the node labels. Jordan equivalence where gi is the geometric multiplicity of eigenvalue 휆i, classes permit identical transforms over graphs of noniden- or the dimension of the kernel of A − 휆iI. The signal tical topologies and allow a basis-invariant characterization space S can be uniquely decomposed by the Jordan of total variation orderings of the spectral components. subspaces (see [13], [14] and Section II). For a graph Methods to exploit these classes to reduce computation time of the transform as well as limitations are discussed. signal s 2 S, the graph Fourier transform (GFT) of [12] is defined as Index Terms—Jordan decomposition, generalized k gi eigenspaces, directed graphs, graph equivalence classes, M M graph isomorphism, signal processing on graphs, networks F : S! Jij i=1 j=1 s ! (s ;:::; s ;:::; s ;:::; s ) ; (1) b11 b1g1 bk1 bkgk I. INTRODUCTION where sij is the (oblique) projection of s onto the Jordan subspace Jij parallel to SnJij. -

EUCLIDEAN DISTANCE MATRIX COMPLETION PROBLEMS June 6
EUCLIDEAN DISTANCE MATRIX COMPLETION PROBLEMS HAW-REN FANG∗ AND DIANNE P. O’LEARY† June 6, 2010 Abstract. A Euclidean distance matrix is one in which the (i, j) entry specifies the squared distance between particle i and particle j. Given a partially-specified symmetric matrix A with zero diagonal, the Euclidean distance matrix completion problem (EDMCP) is to determine the unspecified entries to make A a Euclidean distance matrix. We survey three different approaches to solving the EDMCP. We advocate expressing the EDMCP as a nonconvex optimization problem using the particle positions as variables and solving using a modified Newton or quasi-Newton method. To avoid local minima, we develop a randomized initial- ization technique that involves a nonlinear version of the classical multidimensional scaling, and a dimensionality relaxation scheme with optional weighting. Our experiments show that the method easily solves the artificial problems introduced by Mor´e and Wu. It also solves the 12 much more difficult protein fragment problems introduced by Hen- drickson, and the 6 larger protein problems introduced by Grooms, Lewis, and Trosset. Key words. distance geometry, Euclidean distance matrices, global optimization, dimensional- ity relaxation, modified Cholesky factorizations, molecular conformation AMS subject classifications. 49M15, 65K05, 90C26, 92E10 1. Introduction. Given the distances between each pair of n particles in Rr, n r, it is easy to determine the relative positions of the particles. In many applications,≥ though, we are given only some of the distances and we would like to determine the missing distances and thus the particle positions. We focus in this paper on algorithms to solve this distance completion problem. -

Dimensionality Reduction
CHAPTER 7 Dimensionality Reduction We saw in Chapter 6 that high-dimensional data has some peculiar characteristics, some of which are counterintuitive. For example, in high dimensions the center of the space is devoid of points, with most of the points being scattered along the surface of the space or in the corners. There is also an apparent proliferation of orthogonal axes. As a consequence high-dimensional data can cause problems for data mining and analysis, although in some cases high-dimensionality can help, for example, for nonlinear classification. Nevertheless, it is important to check whether the dimensionality can be reduced while preserving the essential properties of the full data matrix. This can aid data visualization as well as data mining. In this chapter we study methods that allow us to obtain optimal lower-dimensional projections of the data. 7.1 BACKGROUND Let the data D consist of n points over d attributes, that is, it is an n × d matrix, given as ⎛ ⎞ X X ··· Xd ⎜ 1 2 ⎟ ⎜ x x ··· x ⎟ ⎜x1 11 12 1d ⎟ ⎜ x x ··· x ⎟ D =⎜x2 21 22 2d ⎟ ⎜ . ⎟ ⎝ . .. ⎠ xn xn1 xn2 ··· xnd T Each point xi = (xi1,xi2,...,xid) is a vector in the ambient d-dimensional vector space spanned by the d standard basis vectors e1,e2,...,ed ,whereei corresponds to the ith attribute Xi . Recall that the standard basis is an orthonormal basis for the data T space, that is, the basis vectors are pairwise orthogonal, ei ej = 0, and have unit length ei = 1. T As such, given any other set of d orthonormal vectors u1,u2,...,ud ,withui uj = 0 T and ui = 1(orui ui = 1), we can re-express each point x as the linear combination x = a1u1 + a2u2 +···+adud (7.1) 183 184 Dimensionality Reduction T where the vector a = (a1,a2,...,ad ) represents the coordinates of x in the new basis. -

Linear Algebra
Linear Algebra July 28, 2006 1 Introduction These notes are intended for use in the warm-up camp for incoming Berkeley Statistics graduate students. Welcome to Cal! We assume that you have taken a linear algebra course before and that most of the material in these notes will be a review of what you already know. If you have never taken such a course before, you are strongly encouraged to do so by taking math 110 (or the honors version of it), or by covering material presented and/or mentioned here on your own. If some of the material is unfamiliar, do not be intimidated! We hope you find these notes helpful! If not, you can consult the references listed at the end, or any other textbooks of your choice for more information or another style of presentation (most of the proofs on linear algebra part have been adopted from Strang, the proof of F-test from Montgomery et al, and the proof of bivariate normal density from Bickel and Doksum). Go Bears! 1 2 Vector Spaces A set V is a vector space over R and its elements are called vectors if there are 2 opera- tions defined on it: 1. Vector addition, that assigns to each pair of vectors v ; v V another vector w V 1 2 2 2 (we write v1 + v2 = w) 2. Scalar multiplication, that assigns to each vector v V and each scalar r R another 2 2 vector w V (we write rv = w) 2 that satisfy the following 8 conditions v ; v ; v V and r ; r R: 8 1 2 3 2 8 1 2 2 1. -

Linear Algebra with Exercises B
Linear Algebra with Exercises B Fall 2017 Kyoto University Ivan Ip These notes summarize the definitions, theorems and some examples discussed in class. Please refer to the class notes and reference books for proofs and more in-depth discussions. Contents 1 Abstract Vector Spaces 1 1.1 Vector Spaces . .1 1.2 Subspaces . .3 1.3 Linearly Independent Sets . .4 1.4 Bases . .5 1.5 Dimensions . .7 1.6 Intersections, Sums and Direct Sums . .9 2 Linear Transformations and Matrices 11 2.1 Linear Transformations . 11 2.2 Injection, Surjection and Isomorphism . 13 2.3 Rank . 14 2.4 Change of Basis . 15 3 Euclidean Space 17 3.1 Inner Product . 17 3.2 Orthogonal Basis . 20 3.3 Orthogonal Projection . 21 i 3.4 Orthogonal Matrix . 24 3.5 Gram-Schmidt Process . 25 3.6 Least Square Approximation . 28 4 Eigenvectors and Eigenvalues 31 4.1 Eigenvectors . 31 4.2 Determinants . 33 4.3 Characteristic polynomial . 36 4.4 Similarity . 38 5 Diagonalization 41 5.1 Diagonalization . 41 5.2 Symmetric Matrices . 44 5.3 Minimal Polynomials . 46 5.4 Jordan Canonical Form . 48 5.5 Positive definite matrix (Optional) . 52 5.6 Singular Value Decomposition (Optional) . 54 A Complex Matrix 59 ii Introduction Real life problems are hard. Linear Algebra is easy (in the mathematical sense). We make linear approximations to real life problems, and reduce the problems to systems of linear equations where we can then use the techniques from Linear Algebra to solve for approximate solutions. Linear Algebra also gives new insights and tools to the original problems. -

Applied Linear Algebra and Differential Equations
Applied Linear Algebra and Differential Equations Lecture notes for MATH 2350 Jeffrey R. Chasnov The Hong Kong University of Science and Technology Department of Mathematics Clear Water Bay, Kowloon Hong Kong Copyright ○c 2017-2019 by Jeffrey Robert Chasnov This work is licensed under the Creative Commons Attribution 3.0 Hong Kong License. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/hk/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Preface What follows are my lecture notes for a mathematics course offered to second-year engineering students at the the Hong Kong University of Science and Technology. Material from our usual courses on linear algebra and differential equations have been combined into a single course (essentially, two half-semester courses) at the request of our Engineering School. I have tried my best to select the most essential and interesting topics from both courses, and to show how knowledge of linear algebra can improve students’ understanding of differential equations. All web surfers are welcome to download these notes and to use the notes and videos freely for teaching and learning. I also have some online courses on Coursera. You can click on the links below to explore these courses. If you want to learn differential equations, have a look at Differential Equations for Engineers If your interests are matrices and elementary linear algebra, try Matrix Algebra for Engineers If you want to learn vector calculus (also known as multivariable calculus, or calcu- lus three), you can sign up for Vector Calculus for Engineers And if your interest is numerical methods, have a go at Numerical Methods for Engineers Jeffrey R. -

Projection Matrices
Projection Matrices Ed Angel Professor of Computer Science, Electrical and Computer Engineering, and Media Arts University of New Mexico Angel: Interactive Computer Graphics 4E © Addison-Wesley 2005 1 Objectives • Derive the projection matrices used for standard OpenGL projections • Introduce oblique projections • Introduce projection normalization Angel: Interactive Computer Graphics 4E © Addison-Wesley 2005 2 Normalization • Rather than derive a different projection matrix for each type of projection, we can convert all projections to orthogonal projections with the default view volume • This strategy allows us to use standard transformations in the pipeline and makes for efficient clipping Angel: Interactive Computer Graphics 4E © Addison-Wesley 2005 3 Pipeline View modelview projection perspective transformation transformation division 4D → 3D nonsingular clipping Hidden surface removal projection 3D → 2D against default cube Angel: Interactive Computer Graphics 4E © Addison-Wesley 2005 4 Notes • We stay in four-dimensional homogeneous coordinates through both the modelview and projection transformations - Both these transformations are nonsingular - Default to identity matrices (orthogonal view) • Normalization lets us clip against simple cube regardless of type of projection • Delay final projection until end - Important for hidden-surface removal to retain depth information as long as possible Angel: Interactive Computer Graphics 4E © Addison-Wesley 2005 5 Orthogonal Normalization glOrtho(left,right,bottom,top,near,far) normalization -

Questions for Linear Algebra
www.YoYoBrain.com - Accelerators for Memory and Learning Questions for Linear Algebra Category: Default - (148 questions) Linear Algebra: Wronskian of a set of Let (y1, y2, ... yn} be a set of functions which function have n-1 derivatives on an interval I. The determinant:| y1, y2, .... yn || y1', y2', ... yn'||.......................|| y1 (n-1 derivative)..... yn (n-1 derivative) | Linear Algebra: Wronskian test for linear Let { y1, y2, .... yn } be a set of nth order independence linear homogeneous differential equations.This set is linearly independent if and only if the Wronskian is not identically equal to zero Linear Algebra: define the dimension of a if w is a basis of vector space then the vector space number of elements in w is dimension Linear Algebra: if A is m x n matrix define row space - subspace of R^n spanned by row and column space of matrix row vectors of Acolum space subspace of R^m spanned by the column vectors of A Linear Algebra: if A is a m x n matrix, then have the same dimension the row space and the column space of A _____ Linear Algebra: if A is a m x n matrix, then have the same dimension the row space and the column space of A _____ Linear Algebra: define the rank of matrix dimension of the row ( or column ) space of a matrix A is called the rank of Arank( A ) Linear Algebra: how to determine the row put matrix in row echelon form and the space of a matrix non-zero rows form the basis of row space of A Linear Algebra: if A is an m x n matrix of rank n x r r, then the dimension of the solution space of Ax=0 is _____ Linear Algebra: define the coordinate Let B = {v1, v2, .. -

Lecture 5: Matrix Multiplication, Cont.; and Random Projections
Stat260/CS294: Randomized Algorithms for Matrices and Data Lecture 5 - 09/18/2013 Lecture 5: Matrix Multiplication, Cont.; and Random Projections Lecturer: Michael Mahoney Scribe: Michael Mahoney Warning: these notes are still very rough. They provide more details on what we discussed in class, but there may still be some errors, incomplete/imprecise statements, etc. in them. 5 Matrix Multiplication, Cont.; and Random Projections Today, we will use the concentration results of the last few classes to go back and make statements about approximating the product of two matrices; and we will also describe an important topic we will spend a great deal more time on, i.e., random projections and Johnson-Lindenstrauss lemmas. Here is the reading for today. • Dasgupta and Gupta, \An elementary proof of a theorem of Johnson and Lindenstrauss" • Appendix of: Drineas, Mahoney, Muthukrishnan, and Sarlos, \Faster Least Squares Approx- imation" • Achlioptas, \Database-friendly random projections: Johnson-Lindenstrauss with binary coins" 5.1 Spectral norm bounds for matrix multiplication Here, we will provide a spectral norm bound for the error of the approximation constructed by the BasicMatrixMultiplication algorithm. Recall that, given as input a m × n matrix A and an n × p matrix B, this algorithm randomly samples c columns of A and the corresponding rows of B to construct a m × c matrix C and a c × p matrix R such that CR ≈ AB, in the sense that some matrix norm jjAB − CRjj is small. The Frobenius norm bound we established before immediately implies a bound for the spectral norm, but in some cases we will need a better bound than can be obtained in this manner. -

Topics in Random Matrix Theory Terence
Topics in random matrix theory Terence Tao Department of Mathematics, UCLA, Los Angeles, CA 90095 E-mail address: [email protected] To Garth Gaudry, who set me on the road; To my family, for their constant support; And to the readers of my blog, for their feedback and contributions. Contents Preface ix Acknowledgments x Chapter 1. Preparatory material 1 x1.1. A review of probability theory 2 x1.2. Stirling's formula 41 x1.3. Eigenvalues and sums of Hermitian matrices 45 Chapter 2. Random matrices 65 x2.1. Concentration of measure 66 x2.2. The central limit theorem 93 x2.3. The operator norm of random matrices 124 x2.4. The semicircular law 159 x2.5. Free probability 183 x2.6. Gaussian ensembles 217 x2.7. The least singular value 246 x2.8. The circular law 263 Chapter 3. Related articles 277 x3.1. Brownian motion and Dyson Brownian motion 278 x3.2. The Golden-Thompson inequality 297 vii viii Contents x3.3. The Dyson and Airy kernels of GUE via semiclassical analysis 305 x3.4. The mesoscopic structure of GUE eigenvalues 313 Bibliography 321 Index 329 Preface In the spring of 2010, I taught a topics graduate course on random matrix theory, the lecture notes of which then formed the basis for this text. This course was inspired by recent developments in the subject, particularly with regard to the rigorous demonstration of universal laws for eigenvalue spacing distributions of Wigner matri- ces (see the recent survey [Gu2009b]). This course does not directly discuss these laws, but instead focuses on more foundational topics in random matrix theory upon which the most recent work has been based. -
![Arxiv:1811.08406V1 [Math.NA]](https://docslib.b-cdn.net/cover/4287/arxiv-1811-08406v1-math-na-2844287.webp)
Arxiv:1811.08406V1 [Math.NA]
MATRI manuscript No. (will be inserted by the editor) Bj¨orck-Pereyra-type methods and total positivity Jos´e-Javier Mart´ınez Received: date / Accepted: date Abstract The approach to solving linear systems with structured matrices by means of the bidiagonal factorization of the inverse of the coefficient matrix is first considered, the starting point being the classical Bj¨orck-Pereyra algo- rithms for Vandermonde systems, published in 1970 and carefully analyzed by Higham in 1987. The work of Higham showed the crucial role of total pos- itivity for obtaining accurate results, which led to the generalization of this approach to totally positive Cauchy, Cauchy-Vandermonde and generalized Vandermonde matrices. Then, the solution of other linear algebra problems (eigenvalue and singular value computation, least squares problems) is addressed, a fundamental tool being the bidiagonal decomposition of the corresponding matrices. This bidi- agonal decomposition is related to the theory of Neville elimination, although for achieving high relative accuracy the algorithm of Neville elimination is not used. Numerical experiments showing the good behaviour of these algorithms when compared with algorithms which ignore the matrix structure are also included. Keywords Bj¨orck-Pereyra algorithm · Structured matrix · Totally positive matrix · Bidiagonal decomposition · High relative accuracy Mathematics Subject Classification (2010) 65F05 · 65F15 · 65F20 · 65F35 · 15A23 · 15B05 · 15B48 arXiv:1811.08406v1 [math.NA] 20 Nov 2018 J.-J. Mart´ınez Departamento de F´ısica y Matem´aticas, Universidad de Alcal´a, Alcal´ade Henares, Madrid 28871, Spain E-mail: [email protected] 2 Jos´e-Javier Mart´ınez 1 Introduction The second edition of the Handbook of Linear Algebra [25], edited by Leslie Hogben, is substantially expanded from the first edition of 2007 and, in connec- tion with our work, it contains a new chapter by Michael Stewart entitled Fast Algorithms for Structured Matrix Computations (chapter 62) [41]. -

Dimensionality Reduction Via Euclidean Distance Embeddings
School of Computer Science and Communication CVAP - Computational Vision and Active Perception Dimensionality Reduction via Euclidean Distance Embeddings Marin Šarić, Carl Henrik Ek and Danica Kragić TRITA-CSC-CV 2011:2 CVAP320 Marin Sari´c,Carlˇ Henrik Ek and Danica Kragi´c Dimensionality Reduction via Euclidean Distance Embeddings Report number: TRITA-CSC-CV 2011:2 CVAP320 Publication date: Jul, 2011 E-mail of author(s): [marins,chek,dani]@csc.kth.se Reports can be ordered from: School of Computer Science and Communication (CSC) Royal Institute of Technology (KTH) SE-100 44 Stockholm SWEDEN telefax: +46 8 790 09 30 http://www.csc.kth.se/ Dimensionality Reduction via Euclidean Distance Embeddings Marin Sari´c,Carlˇ Henrik Ek and Danica Kragi´c Centre for Autonomous Systems Computational Vision and Active Perception Lab School of Computer Science and Communication KTH, Stockholm, Sweden [marins,chek,dani]@csc.kth.se Contents 1 Introduction2 2 The Geometry of Data3 D 2.1 The input space R : the geometry of observed data.............3 2.2 The configuration region M ...........................4 2.3 The use of Euclidean distance in the input space as a measure of dissimilarity5 q 2.4 Distance-isometric output space R .......................6 3 The Sample Space of a Data Matrix7 3.1 Centering a dataset through projections on the equiangular vector.....8 4 Multidimensional scaling - a globally distance isometric embedding 10 4.1 The relationship between the Euclidean distance matrix and the kernel matrix 11 4.1.1 Generalizing to Mercer kernels..................... 13 4.1.2 Generalizing to any metric....................... 13 4.2 Obtaining output coordinates from an EDM.................