D1ay00963j1.Pdf
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Vidyasagar University Midnapore 721 102
VIDYASAGAR UNIVERSITY MIDNAPORE 721 102 The resolutions of an Emergency Meeting of the Seventh Executive Council of Vidyasagar University dated 04.12.2014. The meeting was attended by the following members : l. Prof. Ranjan Chakrabarti, the Vice-Chancellor. 2. Prof. Ujjayan Bhattacharya. 3. Prof. Bidhan Chandra Patra. 4. Prof. Rajat Acharyya. 5. Dr. Asish Kumar Gupta. 6. Dr. Utpal Kumar Utthasani. 7. Dr. Asim Kumar Bera. 8. Prof. Samir Ghosh. 9. Shri Biswapati Jana. 10. Dr. Joydeb Sasmal. Prof. Ranjan Chakrabarti, the Vice-Chancellor presided over the meeting and Dr. J. K. Nandi, the Registrar(Actg.) acted as the Secretary. Agenda (a) To finalize the matter regarding nominations of two University Nominees in each of the Governing Body of the Undergraduate Colleges, affiliated to Vidyasagar University, for re-constitution of the Governing Body of the Colleges. Resolutions After thorough discussion on the matter relating to nomination of University nominees to the Goveming Body of the Undergraduate Colleges affiliated to Vidyasagar University, it was resolved that two members in each of the Goveming Body of the colleges be nominated as University Nominees for re constitution of the new Goveming Body of the following colleges, specified against each of college, in accordance with the clause (j) of provision 105 of the Vidyasagar University First Statutes, 1983 (amended up;ot 1997) Sl. Name of the Colleges Name of the University Nominees. No. 01. Bajkul Milani Malzavidyalaya, Professor Tapan Kumar Pattanayak, Tamralipta Kismat Bajkul. Mahavidyalaya. Prof Vivekananda Maihf, Khejuri Colle~e. 02. Blzatter College, Dantan. Prof. (Ms.) Sucita Sen, Belda Colle~e. Prof. Sujit Mandai, Kltaragpur College. -

20200302 Vu Ce Gd Piii Cl
Office of the Controller of Examinations VIDYASAGAR UNIVERSITY Midnapore - 721 102 Phone: 03222: 275-441/261-144/276-554-555 Extn.: 419/418/416/451/483/500/531 ............... Ref. No: VU/CE/GD/Partll/CL/2390/2020 Dated: March 02, 2020 Provisional List of centre for B.A. /B.Sc./ B.Com: Honours (Annual Pattern) Part: ~II Examination for the year 2020 .... NEW SYLLABUS . ' SI. Name of the Examination Centre Allotment to the Examination Centre No Colleges to appear No. of candidates Honours H.,~. B.Sc. B.Com Totai 01 Bajkul Milani Mahavidyalaya PK College 792 97 .108 997 Swarnamoyee J Mahavidyalaya YS Palpara Mahavidyalaya 02 Be Ida College Bhattar College, Narayangarh Govt. College, Govt. 391 01 02 394 Gen. Degree College, Dantan 11 03 Bhattar College Belda College 343 43 05 391 04 D<!bra Thana SKS Mahavidyalaya Sabang SK Mahavidyalaya, Siddhinath 343 15 12 370 Mahavidyalaya - 05 Egra SSB College Govt. Gen. Degree College, Mohanpur Ramnagar 301 16 51 368 College 06 Garhbeta College Chandrakona V Mahavidyalaya 523 20 0 543 Gourav Guin Memorial College Paramedical College, r;>urgapur SBSS Mahavidyalaya 07 RS Mahavidyalaya, Ghatal Chaipat SPBS Mahavidyalaya 472 19 0 491 Sukumar Sengupta Mahavidyalaya 08 l·laldia Govt. College Mahishadal Raj College 393 53 45 491 11 ;' 11111111 CJ" ciating) Controller Page ... 1 (Officiating) Vidyasagar University Midnapore-721102, W .B. Downloaded from Vidyasagar University; Copyright (c) Vidyasagar University http://ecircular.vidyasagar.ac.in/DownloadEcircular.aspx?RefId=202003027692 Office of the Controller of Examinations VIDYASAGAR UNIVERSITY Midnapore - 721 102 Phone: 03222: 275-441/261-144/276-554-555 Extn.: 419/418/416/451/483/500/531 .................................................................................................................................................................................................................................................................... -

DBT Star College Scheme, Govt. of India Sponsored Summer School
DBT Star College Scheme, Govt. of India Sponsored Summer School on Advanced Studies on Zoology in The Perception of Current Development Organized by Department of Zoology(UG &PG) Jhargram Raj College Mission & Vision. Jhargram Raj College, being a Government institution, offers quality education programmes at UG as well as a few PG levels under Vidyasagar University and plays an important role in spreading education in the so called backward ‘junglemahal’ area. Many students of this college come from extremely poor families and under-privileged sections of the society from remote rural areas and, are often the first-generation learners. Their requirements are quite different from that of the students belonging to middle class and comparatively well-to-do urban families. To achieve what is required of it, Jhargram Raj College functions accordingly to impart good and quality education in particular and contribute towards social welfare in general. The college has received the prestigious DBT Star scheme to enhance the quality of the learning and teaching process to stimulate original thinking through ‘hands–on’ exposure to experimental work and participation in summer schools. The DBT star scheme promotes networking and strengthens ties with other institutions and laboratories. Department of Zoology, Jhargram Raj College is not an exception and provides an exposure to the students to get the benefit of different subject experts from different colleges, institutes & universities through on line lectures and demonstration of several parts of the syllabi. This will fulfill the completion of least covered part of the syllabi by experts from different academicians including JRC staffs. Course design: The entire course is divided into 6 modules of which 4 are theoretical aspect and the rest 2 will cover the practical area as follows: Duration & Class time: 10 days (14.6.21 to 24.6.21- excluding 15.06.2021), from 11 AM to 2 PM per day; may be preponed or postponed for half an hour, according to situation & availability of Subject experts. -

Prospectus 2021-22
PROSPECTUS 2021-22 VIDYASAGAR UNIVERSITY MIDNAPORE -721 102, WEST BENGAL, INDIA Downloaded from Vidyasagar University; Copyright (c) Vidyasagar University http://ecircular.vidyasagar.ac.in/DownloadEcircular.aspx?RefId=202108298808 From the desk of Vice-Chancellor Let me take this opportunity to welcome you to Vidyasagar University. Created as a State University in 1981, the fledgeling institution flourished rapidly. Having begun on a modest scale in 1985-86, with six post-graduate departments, the University now conducts studies in frontier and interdisciplinary areas of science, social science, humanities and commerce, spread across twenty seven PG departments. The University is proud of its highly qualified and dedicated faculty members, motivated students and research scholars, and adequately equipped supporting and administrative staff who are capable of assimilating the forces of modernization and globalization in the field of higher education. The University is named after Pandit Ishwar Chandra Vidyasagar, the legendary 19th century pioneering social reformer, social activist, litterateur who was dedicated to the cause of the upliftment of women. Since its inception the University has committed itself to the goal of teaching and learning in higher education, ensuring quality education and providing research and extra-curricular activities in the best possible manner in order to spread the benefit of higher education in a location which is relatively backward but which has a glorious history of playing a lead role in the freedom struggle of India. Within the last nine years of its existence, the University has registered its growth both qualitatively and quantitatively. Post- Graduate studies are conducted not only in 27 departments of the University but also in 35 colleges /institutions as well. -

3 Days International Seminar Programme Schedule 2019
Three Days International Seminar on “Environmental History and Sustainability: The Black-White Journey of Sustainable Development in Reality and Education” Organized by Departments of History (UG & PG) & Geography (UG & PG) Bajkul Milani Mahavidyalaya Kismat Bajkul, Purba Medinipur-721655 In Collaboration with World History Research Academy Held on 5th -7th February, 2019 Programme Schedule Day-1: 5th February, 2019 (Tuesday) Time/ Activities/ Person/ Personalities Place/ Venue Duration Functions Frontal Corridor, 09:00 am- Registration & Dept. of - 10:30am Breakfast Geography (UG & PG), BMM Inaugural Session (10:30am – 12:30pm) Invocation Teacher & Students Lighting of lamp Dignitaries Mr. Ardhendu Maity, GB Welcome Address President, BMM & MLA- by Chief Patron Advocate, Bhagwanpur Assembly Constituency Swami 10:30 am – Mr. Rabin Chandra Mandal Vivekananda 12:00 noon Donor Member of GB, BMM , Address by Patron Seminar Hall Member of Alumni Member Association, BMM & Social Worker Dr. P.K.Dandapath, TIC & Address by Chairman, Seminar Chairman Organizing Committee Professor Dr. Kamal Uddin Address by Chief Ahamed, Vice Chancellor, Guest Sher E Bangla Agricultural University, Bangladesh Address about the Mr. G.P.Kar, Secretary, Seminar by Seminar Organizing Organizing Committee Secretary Dr. Pikash Pratim Maity, Principal, Haldia Institute of Health Sciences Mr. Pankaj Konar, BDO, Bhagwanpur-I, Purba Medinipur, West Bengal Dr. Asis Kumar De, Associate Address by Guests of Professor & Head, Honour Department of English (UG and PG), Mahishadal Raj College (NAAC Accredited ‘A’ Grade College) Dr. Anindya Kishor Bhaumik, Ex. Principal, BMM & TIC, Swarnamoyee Jogendranath Mahavidyalaya Address & Vote of Mr. Rabin Das, Convenor Thanks Tea Break (12:30 pm – 12:40pm) 1st Technical Session: 12:40pm – 1:30pm Dr. -

PHATIK GHOSH.Pdf
M I Curriculum Vitae D N Name : DR. PHATIK CHAND GHOSH Designation : Associate Professor A Department of Bengali Midnapore College (Autonomous) P Midnapore- 721101, W.B O Education Qualification : M.A, M.Phil, Ph. D. R Permanent Address : Raghunathpur(Behind Jhargram Nursing Home),Jhargram 701507. E Contact Number : +919832719349/9434481610 Email Id : [email protected] Date Of Joining : 29/03/2007(Mahisadal Raj College) C O List of research papers published : L Ashprishata : Rabindra Natake, Erina, Jhargram, 14-18, Aug 2005 Amarendra Ganaier Kabyanaty Guchha : Punarnaba, Chirantani Kolkata, 9-13, June 2006amader L Satinath Charcha Prasanga Grantha, Divaratrir Kabya, Kolkata 554-585, July-Dec 2006, 2229- 5763 E Nijaswa Shabder Vitar : Kabi Tapan Chakraborty, Sahityer Addy Jhargram, 113-117, G 2008jalpaigurir Kobita : Anchalikatar Sima Chharie, Divaratrir Kabya, Kolkata, 131-176, Jan-April 2009, 2229-5763 E Maniker Uttar Kaler Galpa : Ekti Parichoy, Divaratrir Kabya, Kolkata, 576-588, July-Dec 2009, 2229-5763 Bokul Kathar Shampa : Oithya O Adhunikatar Udvas, Ebong Mushaera, Kolkata, 104-112, Boisakh- A Asar 1417 Nakshal Andolon : Duti Upanyas : Kinnar Roy,Purba Medinipur, 269-292, Sept 2012, 2229-6344 U Nive Jaoa Jumrah, Antarjatik Pathshala, Ghatal , 43-45, Apr-Jun 2013, 2230-9594 Pratibadi Bangla Chhoto Galpa, Purbadesh Galpapatra, Asam, 105-119 , Sharad 1409 T Biswayan O Loksanskriti : Ekti Vabna, Roddur, Kgp,43-49, April 2014 Ashapurna : Satyabati Trilogy :Eisab Purusera, Patachitra, Mahishadal Raj College, 210-229, -

THE WEST BENGAL COLLEGE SERVICE COMMISSION Vacancy Status (Tentative) for the Posts of Assistant Professor in Government-Aided Colleges of West Bengal (Advt
THE WEST BENGAL COLLEGE SERVICE COMMISSION Vacancy Status (Tentative) for the Posts of Assistant Professor in Government-aided Colleges of West Bengal (Advt. No. 1/2018) Bengali UR OBC-A OBC-B SC ST PWD 43 13 1 30 25 6 Sl No College University UR 1 Bankura Zilla Saradamoni Mahila Mahavidyalaya 2 Khatra Adibasi Mahavidyalaya. 3 Panchmura Mahavidyalaya. BANKURA UNIVERSITY 4 Pandit Raghunath Murmu Smriti Mahavidyalaya.(1986) 5 Saltora Netaji Centenary College 6 Sonamukhi College 7 Hiralal Bhakat College 8 Kabi Joydeb Mahavidyalaya 9 Kandra Radhakanta Kundu Mahavidyalaya BURDWAN UNIVERSITY 10 Mankar College 11 Netaji Mahavidyalaya 12 New Alipore College CALCUTTA UNIVERSITY 13 Balurghat Mahila Mahavidyalaya 14 Chanchal College 15 Gangarampur College 16 Harishchandrapur College GOUR BANGA UNIVERSITY 17 Kaliyaganj College 18 Malda College 19 Malda Women's College 20 Pakuahat Degree College 21 Jangipur College 22 Krishnath College 23 Lalgola College KALYANI UNIVERSITY 24 Sewnarayan Rameswar Fatepuria College 25 Srikrishna College 26 Michael Madhusudan Memorial College KAZI NAZRUL UNIVERSITY (ASANSOL) 27 Alipurduar College 28 Falakata College 29 Ghoshpukur College NORTH BENGAL UNIVERSITY 30 Siliguri College 31 Vivekananda College, Alipurduar 32 Mahatma Gandhi College SIDHO KANHO BIRSHA UNIVERSITY 33 Panchakot Mahavidyalaya 34 Bhatter College, Dantan 35 Bhatter College, Dantan 36 Debra Thana Sahid Kshudiram Smriti Mahavidyalaya VIDYASAGAR UNIVERSITY 37 Hijli College 38 Mahishadal Raj College 39 Vivekananda Satavarshiki Mahavidyalaya 40 Dinabandhu -
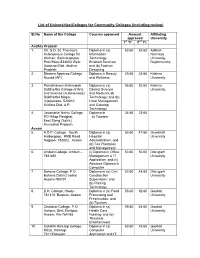
Community Colleges (Including Review)
List of Universities/Colleges for Community Colleges (including review) Sl.No Name of the College Courses approved Amount Affiliating approved University 1st Yr. 2nd Yr. Andhra Pradesh 1. Ch. S.D. St. Theresa’s Diploma in (a) 60.60 53.60 Adikavi Autonomous College for Information Nannaya Women, Sanivarapupet Technology University, Post Eluru-534003 West Enabled Services; Rajahmundry Godavari Dist. Andhra and (b) Fashion Pradesh Designing 2. Dharma Apparao College, Diploma in Beauty 25.65 23.65 Krishna Nuzvid (AP) and Wellness University 3. Parvathaneni Brahmaiah Diploma in (a) 55.60 52.60 Krishna Siddhartha College of Arts Clinical Science University and Science (Autonomous), and Medical Lab Siddhartha Nagar, Technology; and (b) Vijayawada- 520010 Hotel Management Krishna Dist, A.P. and Catering Technology 4. Jawaharlal Nehru College, Diploma in 24.65 23.65 PO Hiltop Pasighat, a) Tourism East Stang District, Arunachal Pradesh Assam 5. A.D.P. College, South Diploma in (a) 50.60 47.60 Guwahati Haiborgaon, RRB Road Hospital University Nagaon- 782002, Assam Administration; and (b) Tea Plantation and Management 6. Amburi College, Amburi – (i) Diploma in Office 60.60 50.60 Dibrugarh 785 680 Management & IT University Application; and (ii) Advance Diploma in Computer 7. Bahona College, P.O Diploma in (a) Civil 52.60 48.60 Dibrugarh Bahona District Jorhat Construction University Assam-785101 Supervision; and (b) Printing Technology 8. B.H. College, Howly- Diploma in (a) Food 55.60 52.60 Gauhati 781316, Barpeta, Assam Processing and University Preservation; and (b) Tourism 9. Chaiduar College, P.O. Diploma in (a) 55.60 46.60 Gauhati Gohpur, Dist: Sonitpur, Health Care University Assam, Pin-784165 Nursing; and (b) Theatre& Entertainment 10. -

MAHISHADAL RAJ COLLEGE (Govt
Serial No. – MAHISHADAL RAJ COLLEGE (Govt. Sponsored) P.O. : MAHISHADAL :: DIST : PURBA MEDINIPUR :: WEST BENGAL APPLICATION FOR ADMISSION TO THE B.A. / B.SC. / B.COM. (HONS. & GEN.) COURSE 1ST YEAR SESSION : 20………. – 20……… Course : B.A. / B.Sc. / B.Com. Honours (Subject …………………………………) / General FOR OFFICE USE Date of admission ………….……... Form issued on ……......…... PASSPORT SIZE Class ………………………….……. PHOTO TO BE PASTED HERE Roll No. ………………………….…. Signature Signature 1. a) Name in full (in Block Letters) : …………………………………………………….…...… Sex – M F b) In Bengali Scripts : ……………………………………………………………………………………....………… c) Address : Vill. : ……………….…………… P.O. : …………..…...………… P.S. : ……….………………….. Dist. : ………………………………………....….…. Contact No. (if any) : ………...………..….…………….. d) Nationality : …………………………………. Religion : ………….………………. Minority : Yes No e) Caste : Gen. S.C. S.T. O.B.C. / Sub caste : …………………..…..…… P.H. : Yes No (Attested copy of Relevant Certificate to be attached) f) Date of Birth : DAY : MONTH : YEAR : 2. a) Father’s Name : ……………………………………………………………………………………………….…… b) Mother’s Name : ………………………………………………………………………………….……………….. c) i) Guardian’s Name ……………………………………………………………...………………………………. ii) Relationship with Guardian : ………………..…………………………… iii) ……………………………….. 3. a) Particulars of H.S. / Equivalent Examination Passed : i) Examination : ………………………… ii) Roll & No. : ……….…………….….….. iii) Year : ……..….….. iv) Name of the Council / Board : ………………………………………………………………………………... v) Name of the School : ………………………………………………………………………………………….. b) Statement of marks -

FIP-NBU-04) (During: February 04, 2021 – March 03, 2021
UGC HUMAN RESOURCE DEVELOPMENT CENTRE UNIVERSITY OF NORTH BENGAL 4th Faculty Induction Programme (FIP-NBU-04) (During: February 04, 2021 – March 03, 2021) “LIST OF SELECTED PARTICIPANTS” Sl. No. / Next Promotion Name Name of the Institute Subject Roll No. Due on HOME UNIVERSITY (N.B.U) 01. Koushik Saha University of North Bengal Geochemistry 15-06-2021 02. Dr. Indrajit Roy Chowdhury Department of Geography And Applied Geography, University of North Bengal Geography 11-08-2021 03. Jamaluddeen Department of Commerce, University of North Bengal Commerce 25-09-2022 04. Kaushani Mondal University of North Bengal English 01-08-2023 05. Sharmishtha Paul University of North Bengal Bengali 13-09-2023 STATE (WEST BENGAL) 06. Dr. Dhritiman Chakraborty RAIGANJ SURENDRANATH MAHAVIDYALAYA ENGLISH 06-08-2020 07. Sk Nasim Ali Nayagram Pandit Raghunath Murmu Government College English 14-08-2020 08. Sudipta Chakraborty Purash Kanpur Haridas Nandi Mahavidyalaya, Howrah, West Bengal Library And Information Science 07-11-2020 09. Nabanita Bhowal Siliguri College Philosophy 12-11-2020 10. Dr. Sucharita Bhattacharyya Barasat College Commerce 18-11-2020 11. Akhil Pandey Midnapore College Microbiology 14-12-2020 12. Prosanta Mandal Sripat Singh College, Jiaganj, Murshidabad Mathematics 05-02-2021 13. Debanjan Seth Purash Kanpur Haridas Nandi Mahavidyalaya English 20-02-2021 14. Samir Kumar Mahato Institute Of Education (P.G.) For Women, Chandernagore, Hooghly. English (Methodology Course) 01-03-2021 15. Bishal Thapa Sonada Degree College English 04-03-2021 16. Debashis Das Shibpur Dinobundhoo Institution (College) Physics 10-03-2021 17. Sushil Kumar Bhuiya Krishnath College Mathematics (Hons) 21-03-2021 18. -

The West Bengal College Service Commission
THE WEST BENGAL COLLEGE SERVICE COMMISSION Vacancy Status for the Posts of Assistant Professor in Government-aided Colleges of West Bengal (Advt. No. 1/2018) The Principal/Vice-Principal/Teacher-in-Charge of the Government-aided College of West Bengal are requested to check the Vacancy status (attached herewith) for the Post of Assistant Professor in the following subject and discrepancy detected, if any, please bring it to the notice of the office of the WBCSC within 5 days. Subject SANSKRIT Date : 30/09/2019 Controller of Examinations THE WEST BENGAL COLLEGE SERVICE COMMISSION Vacancy Status for the Posts of Assistant Professor in Government-aided Colleges of West Bengal (Advt. No. 1/2018) SANSKRIT Sl No. College University UR 1 Hiralal Bhakat College BURDWAN UNIVERSITY 2 M.U.C.Women's College 3 Maharani Kasiswari College 4 Raja Peary Mohan College 5 Ramakrishna Mission Residential College 6 Sadhan Chandra Mahavidyalaya,Harindanga, Falta CALCUTTA UNIVERSITY 7 Sovarani Memorial College 8 Swami Niswambala Nanda Girls' College 9 Udaynarayanpur Madhabilata Mahavidyalaya 10 Vidyasagar College 11 Coochbehar College CPB UNIVERSITY 12 Kaliachak College GOUR BANGA UNIVERSITY 13 Chakdaha College KALYANI UNIVERSITY 14 Srikrishna College 15 Santaldih College SIDHO KANHO BIRSHA UNIVERSITY 16 Bhatter College, Dantan VIDYASAGAR UNIVERSITY 17 Narajole Raj College 18 Acharya Prafulla Chandra College 19 Rishi Bankim Chandra College for Women WEST BENGAL STATE UNIVERSITY 20 Saheed Nurul Islam Mahavidyalaya Sl No. College University OBC-A 1 Onda Thana Mahavidyalaya BANKURA UNIVERSITY 2 Abhedananda Mahavidyalaya 3 Kalna College BURDWAN UNIVERSITY 4 Raja Ram Mohon Roy Mahavidyalaya 5 Narasinha Dutt College 6 Ramakrishna Mission Vidyamandira CALCUTTA UNIVERSITY 7 Serampore Girls' College 8 Asansol Girls' College KAZI NAZRUL UNIVERSITY (ASANSOL) 9 Belda College 10 Mahishadal Raj College VIDYASAGAR UNIVERSITY 11 Sabang Sajanikanta Mahavidyalaya 12 Vivekananda Satavarshiki Mahavidyalaya 13 Dinabandhu Mahvidyalaya (Bongaon) WEST BENGAL STATE UNIVERSITY Sl No. -

Volume 1, Number 1, 2015
The Golden Line A Magazine of English Literature Online version available at www.goldenline.bcdedu.net Inaugural Issue Volume 1, Number 1, 2015 Published by The Department of English Bhatter College, Dantan P.O. Dantan, Dist. Paschim Medinipur West Bengal, India. PIN 721426 Phone: 03229-253238, Fax: 03229-253905 Website: www.bhattercollege.ac.in Email: [email protected] The Golden Line: A Magazine on English Literature Online version available at www.goldenline.bcdedu.net ISSN 2395-1583 (Print) ISSN 2395-1591 (Online) Inaugural Issue Volume 1, Number 1, 2015 Cover Design: The cover includes a painting “near the river” (size 100cm x 120cm) by the artist Edith Spira, Vienna, Austria. Published by The Department of English Bhatter College, Dantan P.O. Dantan, Dist. Paschim Medinipur West Bengal, India. PIN 721426 Phone: 03229-253238, Fax: 03229-253905 Website: www.bhattercollege.ac.in Email: [email protected] © Bhatter College, Dantan Patron Sri Bikram Chandra Pradhan Hon’ble President of the Governing Body, Bhatter College Chief Advisor Pabitra Kumar Mishra Principal, Bhatter College Advisory Board Amitabh Vikram Dwivedi Assistant Professor, Shri Mata Vaishno Devi University, Jammu & Kashmir, India. Indranil Acharya Associate Professor, Vidyasagar University, West Bengal, India. Krishna KBS Assistant Professor in English, Central University of Himachal Pradesh, Dharamshala. Subhajit Sen Gupta Associate Professor, Department of English, Burdwan University. Editor Tarun Tapas Mukherjee Assistant Professor, Department of English, Bhatter College. Editorial Board Santideb Das Guest Lecturer, Department of English, Bhatter College Payel Chakraborty Guest Lecturer, Department of English, Bhatter College Mir Mahammad Ali Guest Lecturer, Department of English, Bhatter College Thakurdas Jana Guest Lecturer, Bhatter College ITI, Bhatter College External Board of Editors Asis De Assistant Professor, Mahishadal Raj College, Vidyasagar University.