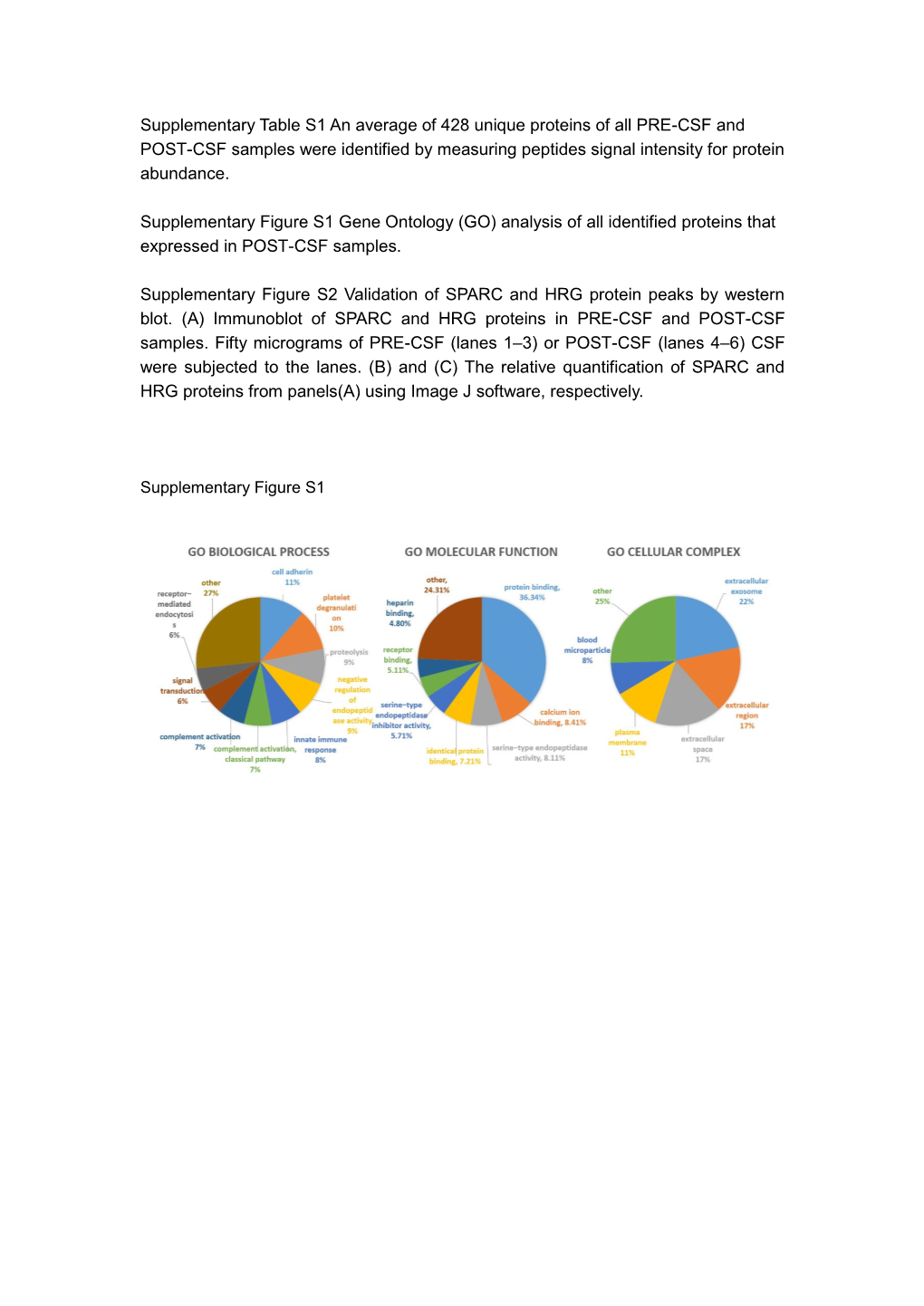
Supplementary Materials
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

How Macrophages Deal with Death
REVIEWS CELL DEATH AND IMMUNITY How macrophages deal with death Greg Lemke Abstract | Tissue macrophages rapidly recognize and engulf apoptotic cells. These events require the display of so- called eat-me signals on the apoptotic cell surface, the most fundamental of which is phosphatidylserine (PtdSer). Externalization of this phospholipid is catalysed by scramblase enzymes, several of which are activated by caspase cleavage. PtdSer is detected both by macrophage receptors that bind to this phospholipid directly and by receptors that bind to a soluble bridging protein that is independently bound to PtdSer. Prominent among the latter receptors are the MER and AXL receptor tyrosine kinases. Eat-me signals also trigger macrophages to engulf virus- infected or metabolically traumatized, but still living, cells, and this ‘murder by phagocytosis’ may be a common phenomenon. Finally , the localized presentation of PtdSer and other eat- me signals on delimited cell surface domains may enable the phagocytic pruning of these ‘locally dead’ domains by macrophages, most notably by microglia of the central nervous system. In long- lived organisms, abundant cell types are often process. Efferocytosis is a remarkably efficient business: short- lived. In the human body, for example, the macrophages can engulf apoptotic cells in less than lifespan of many white blood cells — including neutro- 10 minutes, and it is therefore difficult experimentally to phils, eosinophils and platelets — is less than 2 weeks. detect free apoptotic cells in vivo, even in tissues where For normal healthy humans, a direct consequence of large numbers are generated7. Many of the molecules this turnover is the routine generation of more than that macrophages and other phagocytes use to recognize 100 billion dead cells each and every day of life1,2. -

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Remote Ischemic Preconditioning (RIPC) Modifies Plasma Proteome in Humans
Remote Ischemic Preconditioning (RIPC) Modifies Plasma Proteome in Humans Michele Hepponstall1,2,3,4, Vera Ignjatovic1,3, Steve Binos4, Paul Monagle1,3, Bryn Jones1,2, Michael H. H. Cheung1,2,3, Yves d’Udekem1,2, Igor E. Konstantinov1,2,3* 1 Haematology Research, Murdoch Childrens Research Institute; Melbourne, Victoria, Australia, 2 Cardiac Surgery Unit and Cardiology, Royal Children’s Hospital; Melbourne, Victoria, Australia, 3 Department of Paediatrics, The University of Melbourne; Melbourne, Victoria, Australia, 4 Bioscience Research Division, Department of Primary Industries, Melbourne, Victoria, Australia Abstract Remote Ischemic Preconditioning (RIPC) induced by brief episodes of ischemia of the limb protects against multi-organ damage by ischemia-reperfusion (IR). Although it has been demonstrated that RIPC affects gene expression, the proteomic response to RIPC has not been determined. This study aimed to examine RIPC induced changes in the plasma proteome. Five healthy adult volunteers had 4 cycles of 5 min ischemia alternating with 5 min reperfusion of the forearm. Blood samples were taken from the ipsilateral arm prior to first ischaemia, immediately after each episode of ischemia as well as, at 15 min and 24 h after the last episode of ischemia. Plasma samples from five individuals were analysed using two complementary techniques. Individual samples were analysed using 2Dimensional Difference in gel electrophoresis (2D DIGE) and mass spectrometry (MS). Pooled samples for each of the time-points underwent trypsin digestion and peptides generated were analysed in triplicate using Liquid Chromatography and MS (LC-MS). Six proteins changed in response to RIPC using 2D DIGE analysis, while 48 proteins were found to be differentially regulated using LC-MS. -

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

Calmodulin and Calmodulin-Dependent Protein Kinase II Inhibit Hormone Secretion in Human Parathyroid Adenoma
31 Calmodulin and calmodulin-dependent protein kinase II inhibit hormone secretion in human parathyroid adenoma Ming Lu1,2,3, Erik Berglund1, Catharina Larsson1,3, Anders Ho¨o¨g4, Lars-Ove Farnebo1 and Robert Bra¨nstro¨m1 1Department of Molecular Medicine and Surgery, Karolinska Institutet, Karolinska University Hospital L1:03, SE-171 76 Stockholm, Sweden 2Department of Geriatric Endocrinology, First Affiliated Hospital of Guangxi Medical University, NanNing, People’s Republic of China 3Center for Molecular Medicine (CMM), Karolinska University Hospital, SE-171 76 Stockholm, Sweden 4Department of Oncology–Pathology, Karolinska Institutet, Karolinska University Hospital, SE-171 76 Stockholm, Sweden (Correspondence should be addressed to M Lu at Department of Molecular Medicine and Surgery, Karolinska Institutet, Karolinska University Hospital; Email: [email protected]) Abstract 2C 2C Intracellular calcium ([Ca ]i) is the most relevant modulator adenoma cells in spite of increased [Ca ]i. The inhibitory C of parathyroid hormone (PTH) secretion. Uniquely, an effect of Ca2 calmodulin on PTH secretion may be due to 2C increase in [Ca ]i results in an inhibition of PTH secretion, the absence of synaptotagmin 1 protein in parathyroid and it probably exerts its function via calcium-binding protein adenomas, as demonstrated by western blot analysis. An pathways. The ubiquitous calcium-binding proteins, calmo- increased extracellular calcium level acutely lowered the dulin and calmodulin-dependent protein kinase II (CaMKII), amount of active phosphorylated CaMKII (pCaMKII) in have well-established roles in regulated exocytosis in neurons adenoma cells in vitro, indicating the physiological importance and neuroendocrine cells. However, their roles in parathyroid of this pathway. Moreover, a negative correlation between the cells and PTH secretion are still unclear. -

Complement Component 4 Genes Contribute Sex-Specific Vulnerability in Diverse Illnesses
bioRxiv preprint doi: https://doi.org/10.1101/761718; this version posted September 9, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Complement component 4 genes contribute sex-specific vulnerability in diverse illnesses Nolan Kamitaki1,2, Aswin Sekar1,2, Robert E. Handsaker1,2, Heather de Rivera1,2, Katherine Tooley1,2, David L. Morris3, Kimberly E. Taylor4, Christopher W. Whelan1,2, Philip Tombleson3, Loes M. Olde Loohuis5,6, Schizophrenia Working Group of the Psychiatric Genomics Consortium7, Michael Boehnke8, Robert P. Kimberly9, Kenneth M. Kaufman10, John B. Harley10, Carl D. Langefeld11, Christine E. Seidman1,12,13, Michele T. Pato14, Carlos N. Pato14, Roel A. Ophoff5,6, Robert R. Graham15, Lindsey A. Criswell4, Timothy J. Vyse3, Steven A. McCarroll1,2 1 Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA 2 Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, Massachusetts 02142, USA 3 Department of Medical and Molecular Genetics, King’s College London, London WC2R 2LS, UK 4 Rosalind Russell / Ephraim P Engleman Rheumatology Research Center, Division of Rheumatology, UCSF School of Medicine, San Francisco, California 94143, USA 5 Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, California 90095, USA 6 Center for Neurobehavioral Genetics, Semel Institute for Neuroscience and Human Behavior, University of California, Los Angeles, California 90095, USA 7 A full list of collaborators is in Supplementary Information. -

Investigation of Candidate Genes and Mechanisms Underlying Obesity
Prashanth et al. BMC Endocrine Disorders (2021) 21:80 https://doi.org/10.1186/s12902-021-00718-5 RESEARCH ARTICLE Open Access Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules G. Prashanth1 , Basavaraj Vastrad2 , Anandkumar Tengli3 , Chanabasayya Vastrad4* and Iranna Kotturshetti5 Abstract Background: Obesity associated type 2 diabetes mellitus is a metabolic disorder ; however, the etiology of obesity associated type 2 diabetes mellitus remains largely unknown. There is an urgent need to further broaden the understanding of the molecular mechanism associated in obesity associated type 2 diabetes mellitus. Methods: To screen the differentially expressed genes (DEGs) that might play essential roles in obesity associated type 2 diabetes mellitus, the publicly available expression profiling by high throughput sequencing data (GSE143319) was downloaded and screened for DEGs. Then, Gene Ontology (GO) and REACTOME pathway enrichment analysis were performed. The protein - protein interaction network, miRNA - target genes regulatory network and TF-target gene regulatory network were constructed and analyzed for identification of hub and target genes. The hub genes were validated by receiver operating characteristic (ROC) curve analysis and RT- PCR analysis. Finally, a molecular docking study was performed on over expressed proteins to predict the target small drug molecules. Results: A total of 820 DEGs were identified between -

Bioinformatic Analyses in Early Host Response To
Schroyen et al. BMC Genomics (2016) 17:196 DOI 10.1186/s12864-016-2547-z RESEARCH ARTICLE Open Access Bioinformatic analyses in early host response to Porcine Reproductive and Respiratory Syndrome virus (PRRSV) reveals pathway differences between pigs with alternate genotypes for a major host response QTL Martine Schroyen1†, Christopher Eisley2†, James E. Koltes3, Eric Fritz-Waters1, Igseo Choi4, Graham S. Plastow5, Leluo Guan5, Paul Stothard5, Hua Bao5, Arun Kommadath5, James M. Reecy1, Joan K. Lunney4, Robert R. R. Rowland6, Jack C. M. Dekkers1 and Christopher K. Tuggle1* Abstract Background: AregiononSus scrofa chromosome 4 (SSC4) surrounding single nucleotide polymorphism (SNP) marker WUR10000125 (WUR) has been reported to be strongly associated with both weight gain and serum viremia in pigs after infection with PRRS virus (PRRSV). A proposed causal mutation in the guanylate binding protein 5 gene (GBP5)is predicted to truncate the encoded protein. To investigate transcriptional differences between WUR genotypes in early host response to PRRSV infection, an RNA-seq experiment was performed on globin depleted whole blood RNA collectedon0,4,7,10and14dayspost-infection(dpi)from eight littermate pairs with one AB (favorable) and one AA (unfavorable) WUR genotype animal per litter. Results: Gene Ontology (GO) enrichment analysis of transcripts that were differentially expressed (DE) between dpi across both genotypes revealed an inflammatory response for all dpi when compared to day 0. However, at the early time points of 4 and 7dpi, several GO terms had higher enrichment scores compared to later dpi, including inflammatory response (p <10-7), specifically regulation of NFkappaB (p < 0.01), cytokine, and chemokine activity (p <0.01).At10and 14dpi, GO term enrichment indicated a switch to DNA damage response, cell cycle checkpoints, and DNA replication. -

Pancancer Progression Human Vjune2017
Gene Symbol Accession Alias/Prev Symbol Official Full Name AAMP NM_001087.3 - angio-associated, migratory cell protein ABI3BP NM_015429.3 NESHBP|TARSH ABI family, member 3 (NESH) binding protein ACHE NM_000665.3 ACEE|ARACHE|N-ACHE|YT acetylcholinesterase ACTG2 NM_001615.3 ACT|ACTA3|ACTE|ACTL3|ACTSG actin, gamma 2, smooth muscle, enteric ACVR1 NM_001105.2 ACTRI|ACVR1A|ACVRLK2|ALK2|FOP|SKR1|TSRI activin A receptor, type I ACVR1C NM_145259.2 ACVRLK7|ALK7 activin A receptor, type IC ACVRL1 NM_000020.1 ACVRLK1|ALK-1|ALK1|HHT|HHT2|ORW2|SKR3|TSR-I activin A receptor type II-like 1 ADAM15 NM_207195.1 MDC15 ADAM metallopeptidase domain 15 ADAM17 NM_003183.4 ADAM18|CD156B|CSVP|NISBD|TACE ADAM metallopeptidase domain 17 ADAM28 NM_014265.4 ADAM 28|ADAM23|MDC-L|MDC-Lm|MDC-Ls|MDCL|eMDC II|eMDCII ADAM metallopeptidase domain 28 ADAM8 NM_001109.4 CD156|MS2 ADAM metallopeptidase domain 8 ADAM9 NM_001005845.1 CORD9|MCMP|MDC9|Mltng ADAM metallopeptidase domain 9 ADAMTS1 NM_006988.3 C3-C5|METH1 ADAM metallopeptidase with thrombospondin type 1 motif, 1 ADAMTS12 NM_030955.2 PRO4389 ADAM metallopeptidase with thrombospondin type 1 motif, 12 ADAMTS8 NM_007037.4 ADAM-TS8|METH2 ADAM metallopeptidase with thrombospondin type 1 motif, 8 ADAP1 NM_006869.2 CENTA1|GCS1L|p42IP4 ArfGAP with dual PH domains 1 ADD1 NM_001119.4 ADDA adducin 1 (alpha) ADM2 NM_001253845.1 AM2|dJ579N16.4 adrenomedullin 2 ADRA2B NM_000682.4 ADRA2L1|ADRA2RL1|ADRARL1|ALPHA2BAR|alpha-2BAR adrenoceptor alpha 2B AEBP1 NM_001129.3 ACLP AE binding protein 1 AGGF1 NM_018046.3 GPATC7|GPATCH7|HSU84971|HUS84971|VG5Q -

Appendix Table A.2.3.1 Full Table of All Chicken Proteins and Human Orthologs Pool Accession Human Human Protein Human Product Cell Angios Log2( Endo Gene Comp
Appendix table A.2.3.1 Full table of all chicken proteins and human orthologs Pool Accession Human Human Protein Human Product Cell AngioS log2( Endo Gene comp. core FC) Specific CIKL F1NWM6 KDR NP_002244 kinase insert domain receptor (a type III receptor tyrosine M 94 4 kinase) CWT Q8AYD0 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CWT Q8AYD0 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1P1Y9 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1P1Y9 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1N871 FLT4 NP_891555 fms-related tyrosine kinase 4 M 86 -1.71 CWT O73739 EDNRA NP_001948 endothelin receptor type A M 81 -8 CIKL O73739 EDNRA NP_001948 endothelin receptor type A M 81 -8 CWT Q4ADW2 PROCR NP_006395 protein C receptor, endothelial M 80 -0.36 CIKL Q4ADW2 PROCR NP_006395 protein C receptor, endothelial M 80 -0.36 CIKL F1NFQ9 TEK NP_000450 TEK tyrosine kinase, endothelial M 77 7.3 specific CWT Q9DGN6 ECE1 NP_001106819 endothelin converting enzyme 1 M 74 -0.31 CIKL Q9DGN6 ECE1 NP_001106819 endothelin converting enzyme 1 M 74 -0.31 CWT F1NIF0 CA9 NP_001207 carbonic anhydrase IX I 74 CIKL F1NIF0 CA9 NP_001207 carbonic anhydrase IX I 74 CWT E1BZU7 AOC3 NP_003725 amine oxidase, copper containing 3 (vascular adhesion protein M 70 1) CIKL E1BZU7 AOC3 NP_003725 amine oxidase, copper containing 3 (vascular adhesion protein M 70 1) CWT O93419 COL18A1 NP_569712 collagen, type XVIII, alpha 1 E 70 -2.13 CIKL O93419 -

Supporting Information for Proteomics DOI 10.1002/Prca.200780101
Supporting Information for Proteomics DOI 10.1002/prca.200780101 Paul Cutler, Emma L. Akuffo, Wanda M. Bodnar, Deborah M. Briggs, John B. Davis, Christine M. Debouck, Steven M. Fox, Rachel A. Gibson, Darren A. Gormley, Joanna D. Holbrook, A. Jacqueline Hunter, Emma E. Kinsey, Rabinder Prinjha, Jill C. Richardson, Allen D. Roses, Marjorie A. Smith, Nikos Tsokanas, David R. Will, Wen Wu, John W. Yates and Israel S. Gloger Proteomic identification and early validation of complement 1 inhibitor and pigment epithelium-derived factor: Two novel biomarkers of Alzheimer’s disease in human plasma ª 2007 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim www.clinical.proteomics-journal.com Supplementary Table 1: Complete list of proteins identified from spots derived from 2D gel analysis of human plasma. Each protein was observed to be in a spot showing altered expression between Alzheimer’s disease and matched control by statistical methods as described in the Methods section. Each protein is identified by the gene description and the HUGO gene symbol. The number of “changing” spots in which this protein was observed is also given. HUGO Human Gene Number of Gene Description Symbol Spots alpha-1-B glycoprotein; A1BG 5 alpha-2-macroglobulin A2M 7 afamin; AFM 1 angiotensinogen (serpin peptidase inhibitor, clade A, member 8) AGT 4 alpha-2-HS-glycoprotein AHSG 3 albumin ALB 90 alpha-1-microglobulin/bikunin precursor; AMBP 1 annexin A1 ANXA1 1 amyloid P component, serum APCS 3 apolipoprotein A-I APOA1 14 apolipoprotein A-IV APOA4 2 apolipoprotein E APOE 2 apolipoprotein