Recent Advanced Compuvng Infrastructure Developments At
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

CESMIX: Center for the Exascale Simulation of Materials in Extreme Environments
CESMIX: Center for the Exascale Simulation of Materials in Extreme Environments Project Overview Youssef Marzouk MIT PSAAP-3 Team 18 August 2020 The CESMIX team • Our team integrates expertise in quantum chemistry, atomistic simulation, materials science; hypersonic flow; validation & uncertainty quantification; numerical algorithms; parallel computing, programming languages, compilers, and software performance engineering 1 Project objectives • Exascale simulation of materials in extreme environments • In particular: ultrahigh temperature ceramics in hypersonic flows – Complex materials, e.g., metal diborides – Extreme aerothermal and chemical loading – Predict materials degradation and damage (oxidation, melting, ablation), capturing the central role of surfaces and interfaces • New predictive simulation paradigms and new CS tools for the exascale 2 Broad relevance • Intense current interest in reentry vehicles and hypersonic flight – A national priority! – Materials technologies are a key limiting factor • Material properties are of cross-cutting importance: – Oxidation rates – Thermo-mechanical properties: thermal expansion, creep, fracture – Melting and ablation – Void formation • New systems being proposed and fabricated (e.g., metallic high-entropy alloys) • May have relevance to materials aging • Yet extreme environments are largely inaccessible in the laboratory – Predictive simulation is an essential path… 3 Demonstration problem: specifics • Aerosurfaces of a hypersonic vehicle… • Hafnium diboride (HfB2) promises necessary temperature -

Solid-State NMR Techniques for the Structural Characterization of Cyclic Aggregates Based on Borane–Phosphane Frustrated Lewis Pairs
molecules Review Solid-State NMR Techniques for the Structural Characterization of Cyclic Aggregates Based on Borane–Phosphane Frustrated Lewis Pairs Robert Knitsch 1, Melanie Brinkkötter 1, Thomas Wiegand 2, Gerald Kehr 3 , Gerhard Erker 3, Michael Ryan Hansen 1 and Hellmut Eckert 1,4,* 1 Institut für Physikalische Chemie, WWU Münster, 48149 Münster, Germany; [email protected] (R.K.); [email protected] (M.B.); [email protected] (M.R.H.) 2 Laboratorium für Physikalische Chemie, ETH Zürich, 8093 Zürich, Switzerland; [email protected] 3 Organisch-Chemisches Institut, WWU Münster, 48149 Münster, Germany; [email protected] (G.K.); [email protected] (G.E.) 4 Instituto de Física de Sao Carlos, Universidad de Sao Paulo, Sao Carlos SP 13566-590, Brazil * Correspondence: [email protected] Academic Editor: Mattias Edén Received: 20 February 2020; Accepted: 17 March 2020; Published: 19 March 2020 Abstract: Modern solid-state NMR techniques offer a wide range of opportunities for the structural characterization of frustrated Lewis pairs (FLPs), their aggregates, and the products of cooperative addition reactions at their two Lewis centers. This information is extremely valuable for materials that elude structural characterization by X-ray diffraction because of their nanocrystalline or amorphous character, (pseudo-)polymorphism, or other types of disordering phenomena inherent in the solid state. Aside from simple chemical shift measurements using single-pulse or cross-polarization/magic-angle spinning NMR detection techniques, the availability of advanced multidimensional and double-resonance NMR methods greatly deepened the informational content of these experiments. In particular, methods quantifying the magnetic dipole–dipole interaction strengths and indirect spin–spin interactions prove useful for the measurement of intermolecular association, connectivity, assessment of FLP–ligand distributions, and the stereochemistry of adducts. -

Popular GPU-Accelerated Applications
LIFE & MATERIALS SCIENCES GPU-ACCELERATED APPLICATIONS | CATALOG | AUG 12 LIFE & MATERIALS SCIENCES APPLICATIONS CATALOG Application Description Supported Features Expected Multi-GPU Release Status Speed Up* Support Bioinformatics BarraCUDA Sequence mapping software Alignment of short sequencing 6-10x Yes Available now reads Version 0.6.2 CUDASW++ Open source software for Smith-Waterman Parallel search of Smith- 10-50x Yes Available now protein database searches on GPUs Waterman database Version 2.0.8 CUSHAW Parallelized short read aligner Parallel, accurate long read 10x Yes Available now aligner - gapped alignments to Version 1.0.40 large genomes CATALOG GPU-BLAST Local search with fast k-tuple heuristic Protein alignment according to 3-4x Single Only Available now blastp, multi cpu threads Version 2.2.26 GPU-HMMER Parallelized local and global search with Parallel local and global search 60-100x Yes Available now profile Hidden Markov models of Hidden Markov Models Version 2.3.2 mCUDA-MEME Ultrafast scalable motif discovery algorithm Scalable motif discovery 4-10x Yes Available now based on MEME algorithm based on MEME Version 3.0.12 MUMmerGPU A high-throughput DNA sequence alignment Aligns multiple query sequences 3-10x Yes Available now LIFE & MATERIALS& LIFE SCIENCES APPLICATIONS program against reference sequence in Version 2 parallel SeqNFind A GPU Accelerated Sequence Analysis Toolset HW & SW for reference 400x Yes Available now assembly, blast, SW, HMM, de novo assembly UGENE Opensource Smith-Waterman for SSE/CUDA, Fast short -
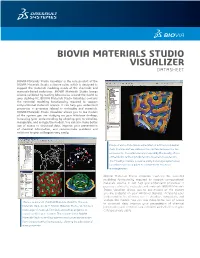
Biovia Materials Studio Visualizer Datasheet
BIOVIA MATERIALS STUDIO VISUALIZER DATASHEET BIOVIA Materials Studio Visualizer is the core product of the BIOVIA Materials Studio software suite, which is designed to support the materials modeling needs of the chemicals and materials-based industries. BIOVIA Materials Studio brings science validated by leading laboratories around the world to your desktop PC. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. You can also make better use of access to structural data, improve your presentation of chemical information, and communicate problems and solutions to your colleagues very easily. Image of early-stage phase segregation in a diblock copolymer melt. The blue surface indicates the interface between the two components. The volume is colormapped by the density of one of the blocks, red being high density, blue being low-density. The MesoDyn module is used to study these large systems over long-times such as required to observe these structural rearrangements. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. -

Download Author Version (PDF)
PCCP Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/pccp Page 1 of 11 PhysicalPlease Chemistry do not adjust Chemical margins Physics PCCP PAPER Effect of nanosize on surface properties of NiO nanoparticles for adsorption of Quinolin-65 ab a a Received 00th January 20xx, Nedal N. Marei, Nashaat N. Nassar* and Gerardo Vitale Accepted 00th January 20xx Using Quinolin-65 (Q-65) as a model-adsorbing compound for polar heavy hydrocarbons, the nanosize effect of NiO Manuscript DOI: 10.1039/x0xx00000x nanoparticles on adsorption of Q-65 was investigated. Different-sized NiO nanoparticles with sizes between 5 and 80 nm were prepared by controlled thermal dehydroxylation of Ni(OH)2. -

Supporting Information
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2015 Supporting Information A Single Crystalline Porphyrinic Titanium Metal−Organic Framework Shuai Yuana†, Tian-Fu Liua†, Dawei Fenga, Jian Tiana, Kecheng Wanga, Junsheng Qina, a a a a b Qiang Zhang , Ying-Pin Chen , Mathieu Bosch , Lanfang Zou , Simon J. Teat, Scott J. c a Dalgarno and Hong-Cai Zhou * a Department of Chemistry, Texas A&M University, College Station, Texas 77842-3012, USA b Advanced Light Source, Lawrence Berkeley National Laboratory Berkeley, CA 94720, USA c Institute of Chemical Sciences, Heriot-Watt University Riccarton, Edinburgh EH14 4AS, U.K. † Equal contribution to this work *To whom correspondence should be addressed. Email: [email protected] Tel: +1 (979) 845-4034; Fax: +1 (979) 845-1595 S1 Contents S1. Ligand Synthesis..............................................................................................................3 S2. Syntheses of PCN-22.......................................................................................................5 S3. X-ray Crystallography .....................................................................................................6 S4. Topological Analyses ......................................................................................................9 S5. N2 Sorption Isotherm .....................................................................................................10 S6. Simulation of the Accessible Surface Area ...................................................................12 -

Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins
Article Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins Marcus Purse 1, Grace Edmund 1, Stephen Hall 1, Brendan Howlin 1,* , Ian Hamerton 2 and Stephen Till 3 1 Department of Chemistry, Faculty of Engineering and Physical Sciences, University of Surrey, Guilford, Surrey GU2 7XH, UK; [email protected] (M.P.); [email protected] (G.E.); [email protected] (S.H.) 2 Bristol Composites Institute (ACCIS), Department of Aerospace Engineering, School of Civil, Aerospace, and Mechanical Engineering, University of Bristol, Bristol BS8 1TR, UK; [email protected] 3 Defence Science and Technology Laboratory, Porton Down, Salisbury SP4 0JQ, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-1483-686-248 Received: 6 March 2019; Accepted: 23 March 2019; Published: 28 March 2019 Abstract: The thermal decomposition of polyphenolic resins was studied by reactive molecular dynamics (RMD) simulation at elevated temperatures. Atomistic models of the polyphenolic resins to be used in the RMD were constructed using an automatic method which calls routines from the software package Materials Studio. In order to validate the models, simulated densities and heat capacities were compared with experimental values. The most suitable combination of force field and thermostat for this system was the Forcite force field with the Nosé–Hoover thermostat, which gave values of heat capacity closest to those of the experimental values. Simulated densities approached a final density of 1.05–1.08 g/cm3 which compared favorably with the experimental values of 1.16–1.21 g/cm3 for phenol-formaldehyde resins. -

TBTK: a Quantum Mechanics Software Development Kit
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Copenhagen University Research Information System TBTK A quantum mechanics software development kit Bjornson, Kristofer Published in: SoftwareX DOI: 10.1016/j.softx.2019.02.005 Publication date: 2019 Document version Publisher's PDF, also known as Version of record Document license: CC BY Citation for published version (APA): Bjornson, K. (2019). TBTK: A quantum mechanics software development kit. SoftwareX, 9, 205-210. https://doi.org/10.1016/j.softx.2019.02.005 Download date: 09. apr.. 2020 SoftwareX 9 (2019) 205–210 Contents lists available at ScienceDirect SoftwareX journal homepage: www.elsevier.com/locate/softx Original software publication TBTK: A quantum mechanics software development kit Kristofer Björnson Niels Bohr Institute, University of Copenhagen, Juliane Maries Vej 30, DK–2100, Copenhagen, Denmark article info a b s t r a c t Article history: TBTK is a software development kit for quantum mechanical calculations and is designed to enable the Received 7 August 2018 development of applications that investigate problems formulated on second-quantized form. It also Received in revised form 21 February 2019 enables method developers to create solvers for tight-binding, DFT, DMFT, quantum transport, etc., that Accepted 22 February 2019 can be easily integrated with each other. Both through the development of completely new solvers, Keywords: as well as front and back ends to already well established packages. TBTK provides data structures Quantum mechanics tailored for second-quantization that will encourage reusability and enable scalability for quantum SDK mechanical calculations. C++ ' 2019 The Author. -

Dmol Guide to Select a Dmol3 Task 1
DMOL3 GUIDE MATERIALS STUDIO 8.0 Copyright Notice ©2014 Dassault Systèmes. All rights reserved. 3DEXPERIENCE, the Compass icon and the 3DS logo, CATIA, SOLIDWORKS, ENOVIA, DELMIA, SIMULIA, GEOVIA, EXALEAD, 3D VIA, BIOVIA and NETVIBES are commercial trademarks or registered trademarks of Dassault Systèmes or its subsidiaries in the U.S. and/or other countries. All other trademarks are owned by their respective owners. Use of any Dassault Systèmes or its subsidiaries trademarks is subject to their express written approval. Acknowledgments and References To print photographs or files of computational results (figures and/or data) obtained using BIOVIA software, acknowledge the source in an appropriate format. For example: "Computational results obtained using software programs from Dassault Systèmes Biovia Corp.. The ab initio calculations were performed with the DMol3 program, and graphical displays generated with Materials Studio." BIOVIA may grant permission to republish or reprint its copyrighted materials. Requests should be submitted to BIOVIA Support, either through electronic mail to [email protected], or in writing to: BIOVIA Support 5005 Wateridge Vista Drive, San Diego, CA 92121 USA Contents DMol3 1 Setting up a molecular dynamics calculation20 Introduction 1 Choosing an ensemble 21 Further Information 1 Defining the time step 21 Tasks in DMol3 2 Defining the thermostat control 21 Energy 3 Constraints during dynamics 21 Setting up the calculation 3 Setting up a transition state calculation 22 Dynamics 4 Which method to use? -

Kepler Gpus and NVIDIA's Life and Material Science
LIFE AND MATERIAL SCIENCES Mark Berger; [email protected] Founded 1993 Invented GPU 1999 – Computer Graphics Visual Computing, Supercomputing, Cloud & Mobile Computing NVIDIA - Core Technologies and Brands GPU Mobile Cloud ® ® GeForce Tegra GRID Quadro® , Tesla® Accelerated Computing Multi-core plus Many-cores GPU Accelerator CPU Optimized for Many Optimized for Parallel Tasks Serial Tasks 3-10X+ Comp Thruput 7X Memory Bandwidth 5x Energy Efficiency How GPU Acceleration Works Application Code Compute-Intensive Functions Rest of Sequential 5% of Code CPU Code GPU CPU + GPUs : Two Year Heart Beat 32 Volta Stacked DRAM 16 Maxwell Unified Virtual Memory 8 Kepler Dynamic Parallelism 4 Fermi 2 FP64 DP GFLOPS GFLOPS per DP Watt 1 Tesla 0.5 CUDA 2008 2010 2012 2014 Kepler Features Make GPU Coding Easier Hyper-Q Dynamic Parallelism Speedup Legacy MPI Apps Less Back-Forth, Simpler Code FERMI 1 Work Queue CPU Fermi GPU CPU Kepler GPU KEPLER 32 Concurrent Work Queues Developer Momentum Continues to Grow 100M 430M CUDA –Capable GPUs CUDA-Capable GPUs 150K 1.6M CUDA Downloads CUDA Downloads 1 50 Supercomputer Supercomputers 60 640 University Courses University Courses 4,000 37,000 Academic Papers Academic Papers 2008 2013 Explosive Growth of GPU Accelerated Apps # of Apps Top Scientific Apps 200 61% Increase Molecular AMBER LAMMPS CHARMM NAMD Dynamics GROMACS DL_POLY 150 Quantum QMCPACK Gaussian 40% Increase Quantum Espresso NWChem Chemistry GAMESS-US VASP CAM-SE 100 Climate & COSMO NIM GEOS-5 Weather WRF Chroma GTS 50 Physics Denovo ENZO GTC MILC ANSYS Mechanical ANSYS Fluent 0 CAE MSC Nastran OpenFOAM 2010 2011 2012 SIMULIA Abaqus LS-DYNA Accelerated, In Development NVIDIA GPU Life Science Focus Molecular Dynamics: All codes are available AMBER, CHARMM, DESMOND, DL_POLY, GROMACS, LAMMPS, NAMD Great multi-GPU performance GPU codes: ACEMD, HOOMD-Blue Focus: scaling to large numbers of GPUs Quantum Chemistry: key codes ported or optimizing Active GPU acceleration projects: VASP, NWChem, Gaussian, GAMESS, ABINIT, Quantum Espresso, BigDFT, CP2K, GPAW, etc. -

An Overview on the Libxc Library of Density Functional Approximations
An overview on the Libxc library of density functional approximations Susi Lehtola Molecular Sciences Software Institute at Virginia Tech 2 June 2021 Outline Why Libxc? Recap on DFT What is Libxc? Using Libxc A look under the hood Wrapup GPAW 2021: Users' and Developers' Meeting Susi Lehtola Why Libxc? 2/28 Why Libxc? There are many approximations for the exchange-correlation functional. But, most programs I ... only implement a handful (sometimes 5, typically 10-15) I ... and the implementations may be buggy / non-standard GPAW 2021: Users' and Developers' Meeting Susi Lehtola Why Libxc? 3/28 Why Libxc, cont'd This leads to issues with reproducibility I chemists and physicists do not traditionally use the same functionals! Outdated(?) stereotype: B3LYP vs PBE I how to reproduce a calculation performed with another code? GPAW 2021: Users' and Developers' Meeting Susi Lehtola Why Libxc? 4/28 Why Libxc, cont'd The issue is compounded by the need for backwards and forwards compatibility: how can one I reproduce old calculations from the literature done with a now-obsolete functional (possibly with a program that is proprietary / no longer available)? I use a newly developed functional in an old program? GPAW 2021: Users' and Developers' Meeting Susi Lehtola Why Libxc? 5/28 Why Libxc, cont'd A standard implementation is beneficial! I no need to keep reinventing (and rebuilding) the wheel I use same collection of density functionals in all programs I new functionals only need to be implemented in one place I broken/buggy functionals only need to be fixed in one place I same implementation can be used across numerical approaches, e.g. -

Electronic Structure Study of Copper-Containing Perovskites
Electronic Structure Study of Copper-containing Perovskites Mark Robert Michel University College London A thesis submitted to University College London in partial fulfilment of the requirements for the degree of Doctor of Philosophy, February 2010. 1 I, Mark Robert Michel, confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. Mark Robert Michel 2 Abstract This thesis concerns the computational study of copper containing perovskites using electronic structure methods. We discuss an extensive set of results obtained using hybrid exchange functionals within Density Functional Theory (DFT), in which we vary systematically the amount of exact (Hartree-Fock, HF) exchange employed. The method has enabled us to obtain accurate results on a range of systems, particularly in materials containing strongly correlated ions, such as Cu2+. This is possible because the HF exchange corrects, at least qualitatively, the spurious self-interaction error present in DFT. The materials investigated include two families of perovskite-structured oxides, of potential interest for technological applications due to the very large dielectric constant or for Multi-Ferroic behaviour. The latter materials exhibit simultaneously ferroelectric and ferromagnetic properties, a rare combination, which is however highly desirable for memory device applications. The results obtained using hybrid exchange functionals are highly encouraging. Initial studies were made on bulk materials such as CaCu3Ti4O12 (CCTO) which is well characterised by experiment. The inclusion of HF exchange improved, in a systematic way, both structural and electronic results with respect to experiment. The confidence gained in the study of known compounds has enabled us to explore new compositions predictively.