Morphometry and Colony Structure of Ants of the Genus Cardiocondyla (Hymenoptera: Formicidae) from Georgia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ORIGINAL ARTICLE a Novel Intracellular Mutualistic Bacterium in the Invasive Ant Cardiocondyla Obscurior
The ISME Journal (2016) 10, 376–388 © 2016 International Society for Microbial Ecology All rights reserved 1751-7362/16 www.nature.com/ismej ORIGINAL ARTICLE A novel intracellular mutualistic bacterium in the invasive ant Cardiocondyla obscurior Antonia Klein1,8, Lukas Schrader1,8, Rosario Gil2, Alejandro Manzano-Marín2, Laura Flórez3, David Wheeler5, John H Werren6, Amparo Latorre2,7, Jürgen Heinze1, Martin Kaltenpoth3,4, Andrés Moya2 and Jan Oettler1 1Institut für Zoologie, Universität Regensburg, Regensburg, Germany; 2Institut Canvanilles de Biodiversitat i Biologia Evolutiva (ICBiBE), Parc Cientific de la Universitat de Valencia, Paterna (Valencia), Spain; 3Max Planck Institute for Chemical Ecology, Jena, Germany; 4Johannes Gutenberg University Mainz, Institute for Zoology, Department for Evolutionary Ecology, Mainz, Germany; 5Institute of Fundamental Sciences, Massey University, Palmerston North, New Zealand; 6Department of Biology, University Rochester, Rochester, NY, USA and 7Área de Genómica y Salud de la Fundación para el Fomento de la Investigación Sanitaria y Biomédica de la Comunitat Valenciana (FISABIO)–Salud Pública, Valencia, Spain The evolution of eukaryotic organisms is often strongly influenced by microbial symbionts that confer novel traits to their hosts. Here we describe the intracellular Enterobacteriaceae symbiont of theinvasiveantCardiocondyla obscurior, ‘Candidatus Westeberhardia cardiocondylae’. Upon metamorphosis, Westeberhardia is found in gut-associated bacteriomes that deteriorate following eclosion. Only queens maintain Westeberhardia in the ovarian nurse cells from where the symbionts are transmitted to late-stage oocytes during nurse cell depletion. Functional analyses of the streamlined genome of Westeberhardia (533 kb, 23.41% GC content) indicate that neither vitamins nor essential amino acids are provided for the host. However, the genome encodes for an almost complete shikimate pathway leading to 4-hydroxyphenylpyruvate, which could be converted into tyrosine by the host. -

Download PDF File
Myrmecological News 23 41-59 Vienna, September 2016 Analyzing large-scale and intranidal phenotype distributions in eusocial Hymenoptera – a taxonomic tool to distinguish intraspecific dimorphism from heterospecificity Bernhard SEIFERT Abstract Ant and termite nests are long-term stable, semi-closed systems constantly producing conspecific worker populations of related individuals over many generations. Accordingly, nests of these eusocial insects, as they are found in nature, offer free of cost an analysis situation that has to be generated in other groups of organisms by controlled rearing experiments. A test system based on analyzing intranidal and large-scale phenotype distributions and comparing the observed distributions with predictions for different scenarios of heterospecificity and intraspecific dimorphism is introduced by a case study on ants. The test system, named DIMORPH test, allows a taxonomist to distinguish if discrete character syndromes represent separate species or an intraspecific phenomenon. One of the most important parameters within the test system is the abundance and distribution of phenotypically mixed nest populations. Five biological explanations are possible for ant nests with a mixture of discrete phenotypes: They may represent (1) geneti- cally determined intraspecific morphs, (2) intraspecific modifications induced by environmental factors, (3) the associ- ation of a temporary social parasite with a host species, (4) the association of a permanent social parasite with a host species, and (5) a parabiotic association of two basically independent (self-sustaining) species. The paper explains the biological background of the scenarios (1) to (5) and presents mathematical models and generalizations from empirical data to predict phenotype distributions for each scenario under variable conditions. -

Hybridization in Ants
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2020 Hybridization in Ants Ian Butler Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons HYBRIDIZATION IN ANTS A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by Ian Butler June 2020 © Copyright by Ian Butler 2020 HYBRIDIZATION IN ANTS Ian Butler, Ph.D. The Rockefeller University 2020 Interspecific hybridization is a relatively common occurrence within all animal groups. Two main factors make hybridization act differently in ants than in other species: eusociality and haplodiploidy. These factors serve to reduce the costs of interspecific hybridization in ants while simultaneously allowing them to take advantage of certain benefits. Eusociality may mitigate the effects of hybridization by allowing hybrids to be shunted into the worker caste, potentially reducing the effects of hybrid sterility. In haplodiploid species, males do not have a father. They instead develop from unfertilized eggs as haploid clones of their mother. This means that interspecifically mated queens do not completely sacrifice reproductive potential even if all hybrids are sterile because they can still produce fertile males. These factors in turn suggest that hybridization should be more common among the social Hymenoptera than other animal groups. Nevertheless, current data suggest that ants hybridize at rates similar to other animal groups, although these data are limited. Furthermore, there is a large amount of overlap between cases of interspecific hybridization and cases of genetic caste determination. A majority of the cases in ants where caste is determined primarily by genotype are associated with hybridization. -

Psyche [March
REVISION OF THE ANT TRIBE CARDIOCONDYLINI (HYMENOPTERA, FORMICIDAE) I. The Genus Prosopidris Wheeler BY JONATHAN REISKIND Biological Laboratories,, Harvard University This is the first of a series of papers revising the tribe Cardio- condylini, a group of small myrmicine ants (up to 3.o mm in length), characterized by the possession of a pedunculate: petiole, a wide postpetiolar dorsum, propodeal spines, and a general lack of pilosity. The tribe currently embraces the genera Cardiocondyla Emery (virtually cosmopolitan, mainly Old World), Xenometra Emery (West Indian) and Prosopidris (Papuasian). Xenometra includes a single poorly known species which is apparently parasitic on Cardio- condyla emeryi (Emery, I917). Cardiocondyla and Prosopidris are free living, and form small inconspicuous colonies: usually in the soil or in rotting logs. In this contribution Wheeler's subgenus Cardiocondyla (Prosopi- dris) is elevated to full generic status. The worker and female, types of its single previously known species, P. sima Wheeler (Philippines), are redescribed and a second species, P. PaDuana (New Guinea), is newly described. The papuana material includes the first known male of the genus, along with females and workers. T'he male is highly ergatoid, like those of some Cardiocondyla species. Measurements and Indices: In order to characterize properly cardio- condyline ants the following measurements and indices (with their abbreviations) will be used in this and succeeding papers: Head Length (HL) Maximum length of head in frontal view from clypeal apex to posterior border of occiput. Head Width (HW)Width of head measured in frontal view immediately behind the eyes. Scape Length (SL) Maximum measurable length of scape, not including its articular condyle. -
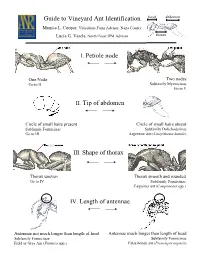
I. Petiole Node II. Tip of Abdomen IV. Length of Antennae Guide to Vineyard Ant Identification III. Shape of Thorax
Guide to Vineyard Ant Identification head abdomen Monica L. Cooper, Viticulture Farm Advisor, Napa County Lucia G. Varela, North Coast IPM Advisor thorax I. Petiole node One Node Two nodes Go to II Subfamily Myrmicinae Go to V II. Tip of abdomen Circle of small hairs present Circle of small hairs absent Subfamily Formicinae Subfamily Dolichoderinae Go to III Argentine Ant (Linepithema humile) III. Shape of thorax Thorax uneven Thorax smooth and rounded Go to IV Subfamily Formicinae Carpenter ant (Camponotus spp.) IV. Length of antennae Antennae not much longer than length of head Antennae much longer than length of head Subfamily Formicinae Subfamily Formicinae Field or Gray Ant (Formica spp.) False honey ant (Prenolepis imparis) head abdomen Petiole with two nodes Subfamily Myrmicinae (V-VIII) thorax V. Dorsal side of Thorax & Antennae One pair of spines on thorax No spines on thorax 12 segmented antennae 10 segmented antennae Go to VI Solenopsis molesta and Solenopsis xyloni VI. Underside of head No brush of bristles Brush of long bristles Go to VII Harvester ants (Pogonomyrmex californicus and P. brevispinosis) VII. Head and Thorax With hairs Without hairs Go to VIII Cardiocondyla mauritanica VIII. Head and Thorax With many parallel furrows Without parallel furrows Profile of thorax rounded Profile of thorax not evenly rounded Pavement ant (Tetramorium “species E”) Pheidole californica Argentine Ant (Linepithema humile), subfamily Dolichoderinae Exotic species 3-4 mm in length Deep brown to light black Move rapidly in distinct trails Feed on honeydew Shallow nests (2 inches from soil surface) Alex Wild Does not bite or sting Carpenter Ant (Camponotus spp.), subfamily Formicinae Large ant: >6 mm in length Dark color with smooth, rounded thorax Workers most active at dusk and night One of most abundant and widespread genera worldwide Generalist scavengers and predators: feed on dead and living insects, nectar, fruit juices and Jack K. -

Institutional Repository - Research Portal Dépôt Institutionnel - Portail De La Recherche
Institutional Repository - Research Portal Dépôt Institutionnel - Portail de la Recherche University of Namurresearchportal.unamur.be RESEARCH OUTPUTS / RÉSULTATS DE RECHERCHE The topology and drivers of ant-symbiont networks across Europe Parmentier, Thomas; DE LAENDER, Frederik; Bonte, Dries Published in: Biological Reviews DOI: Author(s)10.1111/brv.12634 - Auteur(s) : Publication date: 2020 Document Version PublicationPeer reviewed date version - Date de publication : Link to publication Citation for pulished version (HARVARD): Parmentier, T, DE LAENDER, F & Bonte, D 2020, 'The topology and drivers of ant-symbiont networks across PermanentEurope', Biologicallink - Permalien Reviews, vol. : 95, no. 6. https://doi.org/10.1111/brv.12634 Rights / License - Licence de droit d’auteur : General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. BibliothèqueDownload date: Universitaire 07. oct.. 2021 Moretus Plantin 1 The topology and drivers of ant–symbiont networks across 2 Europe 3 4 Thomas Parmentier1,2,*, Frederik de Laender2,† and Dries Bonte1,† 5 6 1Terrestrial Ecology Unit (TEREC), Department of Biology, Ghent University, K.L. -

Recent Human History Governs Global Ant Invasion Dynamics
ARTICLES PUBLISHED: 22 JUNE 2017 | VOLUME: 1 | ARTICLE NUMBER: 0184 Recent human history governs global ant invasion dynamics Cleo Bertelsmeier1*, Sébastien Ollier2, Andrew Liebhold3 and Laurent Keller1* Human trade and travel are breaking down biogeographic barriers, resulting in shifts in the geographical distribution of organ- isms, yet it remains largely unknown whether different alien species generally follow similar spatiotemporal colonization patterns and how such patterns are driven by trends in global trade. Here, we analyse the global distribution of 241 alien ant species and show that these species comprise four distinct groups that inherently differ in their worldwide distribution from that of native species. The global spread of these four distinct species groups has been greatly, but differentially, influenced by major events in recent human history, in particular historical waves of globalization (approximately 1850–1914 and 1960 to present), world wars and global recessions. Species in these four groups also differ in six important morphological and life- history traits and their degree of invasiveness. Combining spatiotemporal distribution data with life-history trait information provides valuable insight into the processes driving biological invasions and facilitates identification of species most likely to become invasive in the future. hallmark of the Anthropocene is range expansion by alien been introduced outside their native range). For each species, we species around the world1, facilitated by the construction of recorded the number of countries where it had established (spatial transport networks and the globalization of trade and labour richness) and estimated spatial diversity taking into account pair- A 2 16 markets since the beginning of the Industrial Revolution . -

Nest Density of Aneuretus Simoni Emery (Sri Lankan Relict Ant) and Stereomyrmex Horni Emery in Three Forest Regions in Western and Southern Sri Lanka
ASIAN MYRMECOLOGY Volume 6, 83–90, 2014 ISSN 1985-1944 © R. K. S. DIA S AN D H. P. G. R. C. RU ch I R ANI Nest density of Aneuretus simoni Emery (Sri Lankan Relict Ant) and Stereomyrmex horni Emery in three forest regions in western and southern Sri Lanka R. K. S. DIA S AN D H. P. G. R. C. RU ch I R ANI Department of Zoology, University of Kelaniya, Sri Lanka Corresponding author’s e-mail: [email protected] ABSTRACT. Aneuretus simoni Emery and Stereomyrmex horni Emery are two endemic and rare ant species in Sri Lanka. The presence or absence and nest density of these species was surveyed to find out if they inhabit locations outside the previously recorded habitats, and to verify their conservation status. Colonies were surveyed by thorough inspection, breaking decaying pieces of wood and the removal of leaf litter in a forest in Western Province (Kalugala Proposed Forest Reserve – KPFR) and two forests in Southern Province (Kuluna Kanda Proposed Forest Reserve – KKPFR and Wilpita “Aranya Kele” in Oliyagan Forest Reserve – WFR). Aneuretus simoni nests were recorded in 7.5% of quadrats, with a nest density of 0.30 nests m−2 at the site of KPFR, whereas this species was more common at KKPFR , occurring in 23.3% of samples with a nest density of 0.93 nests m−2. At WFR site, A. simoni nests were found in 6.7% of samples at a nest density of 0.27 nests m−2. Stereomyrmex horni Emery was found in 15% of quadrats at 0.6 nests m−2 at KPFR and 3% of quadrats at 0.13 nests m−2 at WFR. -

Of Hungary: Survey of Ant Species with an Annotated Synonymic Inventory
insects Article The Myrmecofauna (Hymenoptera: Formicidae) of Hungary: Survey of Ant Species with an Annotated Synonymic Inventory Sándor Cs˝osz 1,2, Ferenc Báthori 2,László Gallé 3,Gábor L˝orinczi 4, István Maák 4,5, András Tartally 6,* , Éva Kovács 7, Anna Ágnes Somogyi 6 and Bálint Markó 8,9 1 MTA-ELTE-MTM Ecology Research Group, Pázmány Péter sétány 1/C, 1117 Budapest, Hungary; [email protected] 2 Evolutionary Ecology Research Group, Centre for Ecological Research, Institute of Ecology and Botany, 2163 Vácrátót, Hungary; [email protected] 3 Department of Ecology and Natural History Collection, University of Szeged, Szeged Boldogasszony sgt. 17., 6722 Szeged, Hungary; [email protected] 4 Department of Ecology, University of Szeged, Közép fasor 52, 6726 Szeged, Hungary; [email protected] (G.L.); [email protected] (I.M.) 5 Museum and Institute of Zoology, Polish Academy of Sciences, ul. Wilcza 64, 00-679 Warsaw, Poland 6 Department of Evolutionary Zoology and Human Biology, University of Debrecen, Egyetem tér 1, 4032 Debrecen, Hungary; [email protected] 7 Kiskunság National Park Directorate, Liszt F. u. 19, 6000 Kecskemét, Hungary; [email protected] 8 Hungarian Department of Biology and Ecology, Babe¸s-BolyaiUniversity, Clinicilor 5-7, 400006 Cluj-Napoca, Romania; [email protected] 9 Centre for Systems Biology, Biodiversity and Bioresources, Babes, -Bolyai University, Clinicilor 5-7, 400006 Cluj-Napoca, Romania * Correspondence: [email protected]; Tel.: +36-52-512-900 (ext. 62349) Simple Summary: Abundance is a hallmark of ants (Hymenoptera: Formicidae). They are exceed- ingly common in both natural and artificial environments and they constitute a conspicuous part Citation: Cs˝osz,S.; Báthori, F.; Gallé, of the terrestrial ecosystem; every 3 to 4 out of 10 kg of insects are given by ants. -

Hymenoptera: Formicidae) 315-321 © Zool.-Bot
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Verhandlungen der Zoologisch-Botanischen Gesellschaft in Wien. Frueher: Verh.des Zoologisch-Botanischen Vereins in Wien. seit 2014 "Acta ZooBot Austria" Jahr/Year: 2018 Band/Volume: 155_1 Autor(en)/Author(s): Wagner Herbert Christian, Seifert Bernhard, Borovsky Roman, Paill Wolfgang Artikel/Article: First insight into the ant diversity of the Vjosa valley, Albania (Hymenoptera: Formicidae) 315-321 © Zool.-Bot. Ges. Österreich, Austria; download unter www.zobodat.at Acta ZooBot Austria 155, 2018, 315–321 First insight into the ant diversity of the Vjosa valley, Albania (Hymenoptera: Formicidae) Herbert Christian Wagner, Bernhard Seifert, Roman Borovsky & Wolfgang Paill Thirty sites at the river Vjosa (Albania) were sampled with 150 pitfall traps in April 2017. The ant catches were determined and are presented. We identified 19 species; two of them show ripicolous tendencies. Nine species are considered to be first records for Albania; this demonstrates the poor investigation status of this territory. WAGNER H.CH., SEIFERT B., BOROVSKY R. & Wolfgang PAILL W., 2018: Erster Ein- blick in die Ameisendiversität des Vjosa-Tales, Albanien (Hymenoptera: Formi- cidae). Dreißig Standorte am Fluss Vjosa (Albanien) wurden im April 2017 mit 150 Barber- fallen beprobt. Die Ameisenfänge wurden determiniert und werden vorgestellt. Wir identifizierten 19 Arten, zwei davon zeigen ripikole Tendenzen. Neun Arten werden als Erstnachweise für Albanien betrachtet, was den schlechten Erforschungsstand dieses Landes demonstriert. Keywords: Vjosa river, floodplain, riparian, ripicol, new records. Introduction The Balkans probably comprise the largest ant diversity of Europe (Borowiec & Salata 2012b, Borowiec 2014, Bračko et al. -

Sexual Cooperation in Two Male-Diphenic Cardiocondyla Species
Sexual cooperation in two male-diphenic Cardiocondyla species DISSERTATION ZUR ERLANGUNG DES DOKTORGRADES DER NATURWISSENSCHAFTEN (DR. RER. NAT.) DER FAKULTÄT FÜR BIOLOGIE UND VORKLINISCHE MEDIZIN DER UNIVERSITÄT REGENSBURG vorgelegt von Marion Füßl aus Ingolstadt im Jahr 2014 Sexual cooperation in two male-diphenic Cardiocondyla species DISSERTATION ZUR ERLANGUNG DES DOKTORGRADES DER NATURWISSENSCHAFTEN (DR. RER. NAT.) DER FAKULTÄT FÜR BIOLOGIE UND VORKLINISCHE MEDIZIN DER UNIVERSITÄT REGENSBURG vorgelegt von Marion Füßl aus Ingolstadt im Jahr 2014 Promotionsgesuch eingereicht am: 19.12.2014 Die Arbeit wurde angeleitet von Dr. A. Schrempf Prüfungsausschuss: Vorsitzender: Prof. Dr. S. Schneuwly 1. Gutachter: Prof. Dr. J. Heinze 2. Gutachter: Prof. Dr. J. Ruther 3. Prüfer: Prof. Dr. C. Oberprieler Unterschrift: If you watch animals objectively for any length of time, you’re driven to the conclusion that their main aim in life is to pass on their genes to the next generation. – David Attenborough – Manuscripts that contributed to this Thesis and Author’s contribution Fuessl M, Santos CG, Bayersdorfer F, Skorupa J, Hartfelder KH, Heinze J, Schrempf A (in preparation) Suppression subtractive hybridization reveals differentially expressed genes in the accessory glands of two ant male phenes with different life histories, (Chapter 2) Author’s contribution: JH, KHH and AS designed and supervised the study. FB gave helpful advice for qPCR. AS contributed to the manuscript. MF and CGS performed the RDA. JS performed the qPCR which was supervised by MF and AS. Bioinformatic analysis to obtain USs were done by CGS and database searches by MF. Dissection, cDNA-synthesis, primer design, sample preparations, evaluation of qPCR results and writing the draft of the manuscript was done by MF. -

Martin Pfeiffer-Habilitation
Structuring of animal communities: Interspecific interactions and habitat selection among ants and small mammals Habilitationsschrift zur Erlangung der Venia Legendi an der Universität Ulm Fakultät für Naturwissenschaften vorgelegt von Dr. Martin Pfeiffer Ulm, Oktober 2007 Year’s end — Still in straw hat And sandals. Basho (1644 -1694) In dieser Arbeit werden die Untersuchungen zur Gemeinschaftsökologie von Ameisen und Kleinsäugern vorgestellt, die ich zwischen 1997 und 2007 durchgeführt oder betreut habe. Ich versichere, dass ich die vorliegende Arbeit ohne fremde Hilfe angefertigt und mich keiner anderen als der ausdrücklich angegebenen Hilfsmittel bedient habe. Martin Pfeiffer Ulm, 30. Oktober 2007 CONTENTS Acknowledgments 1 1. Disentangling life histories, organization, and functions in animal communities of tropical rainforests and arid areas – an overview 3 2. Internet-based ant taxonomy and biodiversity informatics 8 3. Ant diversity gradients and faunistic inventory 14 4. Null model studies of interspecific interactions: community structure of Malaysian ants 18 5. The Sarawak soil ant project: Niches, trophic levels, and community patterns in rainforest ants 21 6. Ant- plant mutualism: Myrmecochory - seed dispersal by ants 24 7. Spatial organization in Bornean small mammal assemblages 27 8. Rainforest logging in Borneo: impacts on non-volant small mammal assemblages 30 References 34 Research articles ordered Research articles belonging to Chapter 3 43 Pfeiffer M, Chimedregzen L, Ulykpan K (2003) Community organization and species richness of ants (Hymenoptera/Formicidae) in Mongolia along an ecological gradient from steppe to Gobi desert. Journal of Biogeography 30:1921-1935 Pfeiffer M, Schultz R, Radchenko A, Yamane S, Woyciechowski M, Ulykpan A, Seifert B (2006) A critical checklist of the ants of Mongolia (Hymenoptera : Formicidae).