In Search of the Tree of Life for Turtles
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Geographic Variation in the Matamata Turtle, Chelus Fimbriatus, with Observations on Its Shell Morphology and Morphometry
n*entilkilt ilil Biok,gr', 1995. l(-l):19: 1995 by CheloninD Research Foundltion Geographic Variation in the Matamata Turtle, Chelus fimbriatus, with Observations on its Shell Morphology and Morphometry MlncBLo R. SANcnnz-Vu,urcnAr, PnrER C.H. PnrrcHARD:, ArrnEro P.rorrLLo-r, aNn Onan J. LINlnBs3 tDepartment of Biological Anthropolog-,- and Anatomy, Duke lJniversin' Medical Cetter. Box 3170, Dtu'hcun, North Carolina277l0 USA IFat 919-684-8034]; 2Florida Audubotr Societ-t, 460 High,n;a,- 436, Suite 200, Casselberry, Florida 32707 USA: iDepartanento de Esttdios Anbientales, llniyersitlad Sinzrin Bolltnt", Caracas ]O80-A, APDO 89OOO l/enerte\a Ansrucr. - A sample of 126 specimens of Chelusftmbriatus was examined for geographic variation and morphology of the shell. A high degree of variation was found in the plastral formula and in the shape and size of the intergular scute. This study suggests that the Amazon population of matamatas is different from the Orinoco population in the following characters: shape ofthe carapace, plastral pigmentation, and coloration on the underside of the neck. Additionatly, a preliminary analysis indicates that the two populations could be separated on the basis of the allometric growth of the carapace in relation to the plastron. Kry Wonus. - Reptilia; Testudinesl Chelidae; Chelus fimbriatus; turtle; geographic variationl allometryl sexual dimorphism; morphology; morphometryl osteology; South America 'Ihe matamata turtle (Chelus fimbricttus) inhabits the scute morpholo..ey. Measured characters (in all cases straight- Amazon, Oyapoque. Essequibo. and Orinoco river systems line) were: maximum carapace len.-uth (CL). cArapace width of northern South America (Iverson. 1986). Despite a mod- at the ler,'el of the sixth marginal scute (CW). -

A New Xinjiangchelyid Turtle from the Middle Jurassic of Xinjiang, China and the Evolution of the Basipterygoid Process in Mesozoic Turtles Rabi Et Al
A new xinjiangchelyid turtle from the Middle Jurassic of Xinjiang, China and the evolution of the basipterygoid process in Mesozoic turtles Rabi et al. Rabi et al. BMC Evolutionary Biology 2013, 13:203 http://www.biomedcentral.com/1471-2148/13/203 Rabi et al. BMC Evolutionary Biology 2013, 13:203 http://www.biomedcentral.com/1471-2148/13/203 RESEARCH ARTICLE Open Access A new xinjiangchelyid turtle from the Middle Jurassic of Xinjiang, China and the evolution of the basipterygoid process in Mesozoic turtles Márton Rabi1,2*, Chang-Fu Zhou3, Oliver Wings4, Sun Ge3 and Walter G Joyce1,5 Abstract Background: Most turtles from the Middle and Late Jurassic of Asia are referred to the newly defined clade Xinjiangchelyidae, a group of mostly shell-based, generalized, small to mid-sized aquatic froms that are widely considered to represent the stem lineage of Cryptodira. Xinjiangchelyids provide us with great insights into the plesiomorphic anatomy of crown-cryptodires, the most diverse group of living turtles, and they are particularly relevant for understanding the origin and early divergence of the primary clades of extant turtles. Results: Exceptionally complete new xinjiangchelyid material from the ?Qigu Formation of the Turpan Basin (Xinjiang Autonomous Province, China) provides new insights into the anatomy of this group and is assigned to Xinjiangchelys wusu n. sp. A phylogenetic analysis places Xinjiangchelys wusu n. sp. in a monophyletic polytomy with other xinjiangchelyids, including Xinjiangchelys junggarensis, X. radiplicatoides, X. levensis and X. latiens. However, the analysis supports the unorthodox, though tentative placement of xinjiangchelyids and sinemydids outside of crown-group Testudines. A particularly interesting new observation is that the skull of this xinjiangchelyid retains such primitive features as a reduced interpterygoid vacuity and basipterygoid processes. -
![Interpreting Character Variation in Turtles: [I]Araripemys Barretoi](https://docslib.b-cdn.net/cover/3241/interpreting-character-variation-in-turtles-i-araripemys-barretoi-123241.webp)
Interpreting Character Variation in Turtles: [I]Araripemys Barretoi
A peer-reviewed version of this preprint was published in PeerJ on 29 September 2020. View the peer-reviewed version (peerj.com/articles/9840), which is the preferred citable publication unless you specifically need to cite this preprint. Limaverde S, Pêgas RV, Damasceno R, Villa C, Oliveira GR, Bonde N, Leal MEC. 2020. Interpreting character variation in turtles: Araripemys barretoi (Pleurodira: Pelomedusoides) from the Araripe Basin, Early Cretaceous of Northeastern Brazil. PeerJ 8:e9840 https://doi.org/10.7717/peerj.9840 Interpreting character variation in turtles: Araripemys barretoi (Pleurodira: Pelomedusoides) from the Araripe Basin, Early Cretaceous of Northeastern Brazil Saulo Limaverde 1 , Rodrigo Vargas Pêgas 2 , Rafael Damasceno 3 , Chiara Villa 4 , Gustavo Oliveira 3 , Niels Bonde 5, 6 , Maria E. C. Leal Corresp. 1, 5 1 Centro de Ciências, Departamento de Geologia, Universidade Federal do Ceará, Fortaleza, Brazil 2 Department of Geology and Paleontology, Museu Nacional/Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 3 Departamento de Biologia, Universidade Federal Rural de Pernambuco, Recife, Brazil 4 Department of Forensic Medicine, Copenhagen University, Copenhagen, Denmark 5 Section Biosystematics, Zoological Museum (SNM, Copenhagen University), Copenhagen, Denmark 6 Fur Museum (Museum Saling), Fur, DK-7884, Denmark Corresponding Author: Maria E. C. Leal Email address: [email protected] The Araripe Basin (Northeastern Brazil) has yielded a rich Cretaceous fossil fauna of both vertebrates and invertebrates found mainly in the Crato and Romualdo Formations, of Aptian and Albian ages respectively. Among the vertebrates, the turtles were proved quite diverse, with several specimens retrieved and five valid species described to this date for the Romualdo Fm. -

Carettochelys Insculpta) in the KIKORI REGION, PAPUA NEW GUINEA
NESTING ECOLOGY, HARVEST AND CONSERVATION OF THE PIG-NOSED TURTLE (Carettochelys insculpta) IN THE KIKORI REGION, PAPUA NEW GUINEA by CARLA CAMILO EISEMBERG DE ALVARENGA B.Sc. (Federal University of Minas Gerais – UFMG) (2004) M.Sc. (National Institute for Amazon Research) (2006) Institute for Applied Ecology University of Canberra Australia A thesis submitted in fulfilment of the requirements of the Degree of Doctor of Philosophy at the University of Canberra. October 2010 Certificate of Authorship of Thesis Except where clearly acknowledged in footnotes, quotations and the bibliography, I certify that I am the sole author of the thesis submitted today, entitled Nesting ecology, harvest and conservation of the Pig-nosed turtle (Carettochelys insculpta) in the Kikori region, Papua New Guinea I further certify that to the best of my knowledge the thesis contains no material previously published or written by another person except where due reference is made in the text of the thesis. The material in the thesis has not been the basis of an award of any other degree or diploma.The thesis complies with University requirements for a thesis as set out in Gold Book Part 7: Examination of Higher Degree by Research Theses Policy, Schedule Two (S2). …………………………………………… Signature of Candidate .......................................................................... Signature of chair of the supervisory panel ………15-Mar-11…………………….. Date Copyright This thesis (© by Carla C. Eisemberg, 2010) may be freely copied or distributed for private and/or commercial use and study. However, no part of this thesis or the information herein may be included in a publication or referred to in a publication without the written consent of Carla C. -
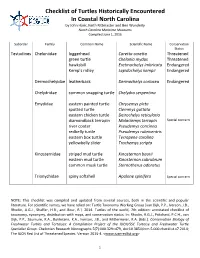
N.C. Turtles Checklist
Checklist of Turtles Historically Encountered In Coastal North Carolina by John Hairr, Keith Rittmaster and Ben Wunderly North Carolina Maritime Museums Compiled June 1, 2016 Suborder Family Common Name Scientific Name Conservation Status Testudines Cheloniidae loggerhead Caretta caretta Threatened green turtle Chelonia mydas Threatened hawksbill Eretmochelys imbricata Endangered Kemp’s ridley Lepidochelys kempii Endangered Dermochelyidae leatherback Dermochelys coriacea Endangered Chelydridae common snapping turtle Chelydra serpentina Emydidae eastern painted turtle Chrysemys picta spotted turtle Clemmys guttata eastern chicken turtle Deirochelys reticularia diamondback terrapin Malaclemys terrapin Special concern river cooter Pseudemys concinna redbelly turtle Pseudemys rubriventris eastern box turtle Terrapene carolina yellowbelly slider Trachemys scripta Kinosternidae striped mud turtle Kinosternon baurii eastern mud turtle Kinosternon subrubrum common musk turtle Sternotherus odoratus Trionychidae spiny softshell Apalone spinifera Special concern NOTE: This checklist was compiled and updated from several sources, both in the scientific and popular literature. For scientific names, we have relied on: Turtle Taxonomy Working Group [van Dijk, P.P., Iverson, J.B., Rhodin, A.G.J., Shaffer, H.B., and Bour, R.]. 2014. Turtles of the world, 7th edition: annotated checklist of taxonomy, synonymy, distribution with maps, and conservation status. In: Rhodin, A.G.J., Pritchard, P.C.H., van Dijk, P.P., Saumure, R.A., Buhlmann, K.A., Iverson, J.B., and Mittermeier, R.A. (Eds.). Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. Chelonian Research Monographs 5(7):000.329–479, doi:10.3854/crm.5.000.checklist.v7.2014; The IUCN Red List of Threatened Species. -

A Phylogenomic Analysis of Turtles ⇑ Nicholas G
Molecular Phylogenetics and Evolution 83 (2015) 250–257 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev A phylogenomic analysis of turtles ⇑ Nicholas G. Crawford a,b,1, James F. Parham c, ,1, Anna B. Sellas a, Brant C. Faircloth d, Travis C. Glenn e, Theodore J. Papenfuss f, James B. Henderson a, Madison H. Hansen a,g, W. Brian Simison a a Center for Comparative Genomics, California Academy of Sciences, 55 Music Concourse Drive, San Francisco, CA 94118, USA b Department of Genetics, University of Pennsylvania, Philadelphia, PA 19104, USA c John D. Cooper Archaeological and Paleontological Center, Department of Geological Sciences, California State University, Fullerton, CA 92834, USA d Department of Biological Sciences, Louisiana State University, Baton Rouge, LA 70803, USA e Department of Environmental Health Science, University of Georgia, Athens, GA 30602, USA f Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA g Mathematical and Computational Biology Department, Harvey Mudd College, 301 Platt Boulevard, Claremont, CA 9171, USA article info abstract Article history: Molecular analyses of turtle relationships have overturned prevailing morphological hypotheses and Received 11 July 2014 prompted the development of a new taxonomy. Here we provide the first genome-scale analysis of turtle Revised 16 October 2014 phylogeny. We sequenced 2381 ultraconserved element (UCE) loci representing a total of 1,718,154 bp of Accepted 28 October 2014 aligned sequence. Our sampling includes 32 turtle taxa representing all 14 recognized turtle families and Available online 4 November 2014 an additional six outgroups. Maximum likelihood, Bayesian, and species tree methods produce a single resolved phylogeny. -

A Case of Inversion and Duplication Involving Constitutive Heterochromatin
Genetics and Molecular Biology, 36, 3, 353-356 (2013) Copyright © 2013, Sociedade Brasileira de Genética. Printed in Brazil www.sbg.org.br Short Communication Cytogenetic comparison of Podocnemis expansa and Podocnemis unifilis: A case of inversion and duplication involving constitutive heterochromatin Ricardo José Gunski1, Isabel Souza Cunha2, Tiago Marafiga Degrandi1, Mario Ledesma3 and Analía Del Valle Garnero1 1Universidade Federal do Pampa, Campus São Gabriel, São Gabriel, RS, Brazil. 2Secretaria Municipal de Meio Ambiente e Turismo, São Desidério, BA, Brazil. 3Parque Ecológico “El Puma”, Candelaria, Argentina. Abstract Podocnemis expansa and P. unifilis present 2n = 28 chromosomes, a diploid number similar to those observed in other species of the genus. The aim of this study was to characterize these two species using conventional staining and differential CBG-, GTG and Ag-NOR banding. We analyzed specimens of P. expansa and P. unifilis from the state of Tocantins (Brazil), in which we found a 2n = 28 and karyotypes differing in the morphology of the 13th pair, which was submetacentric in P. expansa and telocentric in P. unifilis. The CBG-banding patterns revealed a heterochromatic block in the short arm of pair 13 of P. expansa and an interstitial one in pair 13 of P. unifilis, suggest- ing a pericentric inversion. Pair 14 of P. unifilis showed an insterstitial band in the long arm that was absent in P. expansa, suggesting a duplication in this region. Ag-NORs were observed in the first chromosome pair of both spe- cies and was associated to a secondary constriction and heterochromatic blocks. Keywords: chromosome banding, Ag-NORs, karyotype, chelonids. -

Dermatemys Mawii (The Hicatee, Tortuga Blanca, Or Central American River Turtle): a Working Bibliography
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/325206006 Dermatemys mawii (The Hicatee, Tortuga Blanca, or Central American River Turtle): A Working Bibliography Article · May 2018 CITATIONS READS 0 521 6 authors, including: Venetia Briggs Sergio C. Gonzalez University of Florida University of Florida 16 PUBLICATIONS 133 CITATIONS 9 PUBLICATIONS 39 CITATIONS SEE PROFILE SEE PROFILE Thomas Rainwater Clemson University 143 PUBLICATIONS 1,827 CITATIONS SEE PROFILE Some of the authors of this publication are also working on these related projects: Crocodiles in the Everglades View project Dissertation: The role of habitat expansion on amphibian community structure View project All content following this page was uploaded by Sergio C. Gonzalez on 17 May 2018. The user has requested enhancement of the downloaded file. 2018 Endangered and ThreatenedCaribbean Species Naturalist of the Caribbean Region Special Issue No. 2 V. Briggs-Gonzalez, S.C. Gonzalez, D. Smith, K. Allen, T.R. Rainwater, and F.J. Mazzotti 2018 CARIBBEAN NATURALIST Special Issue No. 2:1–22 Dermatemys mawii (The Hicatee, Tortuga Blanca, or Central American River Turtle): A Working Bibliography Venetia Briggs-Gonzalez1,*, Sergio C. Gonzalez1, Dustin Smith2, Kyle Allen1, Thomas R. Rainwater3, and Frank J. Mazzotti1 Abstract - Dermatemys mawii (Central American River Turtle), locally known in Belize as the “Hicatee” and in Guatemala and Mexico as Tortuga Blanca, is a large, highly aquatic freshwater turtle that has been extirpated from much of its historical range of southern Mex- ico, northern Guatemala, and lowland Belize. Throughout its restricted range, Dermatemys has been intensely harvested for its meat and eggs and sold in local markets. -

Membros Da Comissão Julgadora Da Dissertação
UNIVERSIDADE DE SÃO PAULO FACULDADE DE FILOSOFIA, CIÊNCIAS E LETRAS DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM BIOLOGIA COMPARADA Evolution of the skull shape in extinct and extant turtles Evolução da forma do crânio em tartarugas extintas e viventes Guilherme Hermanson Souza Dissertação apresentada à Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto da Universidade de São Paulo, como parte das exigências para obtenção do título de Mestre em Ciências, obtido no Programa de Pós- Graduação em Biologia Comparada Ribeirão Preto - SP 2021 UNIVERSIDADE DE SÃO PAULO FACULDADE DE FILOSOFIA, CIÊNCIAS E LETRAS DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM BIOLOGIA COMPARADA Evolution of the skull shape in extinct and extant turtles Evolução da forma do crânio em tartarugas extintas e viventes Guilherme Hermanson Souza Dissertação apresentada à Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto da Universidade de São Paulo, como parte das exigências para obtenção do título de Mestre em Ciências, obtido no Programa de Pós- Graduação em Biologia Comparada. Orientador: Prof. Dr. Max Cardoso Langer Ribeirão Preto - SP 2021 Autorizo a reprodução e divulgação total ou parcial deste trabalho, por qualquer meio convencional ou eletrônico, para fins de estudo e pesquisa, desde que citada a fonte. I authorise the reproduction and total or partial disclosure of this work, via any conventional or electronic medium, for aims of study and research, with the condition that the source is cited. FICHA CATALOGRÁFICA Hermanson, Guilherme Evolution of the skull shape in extinct and extant turtles, 2021. 132 páginas. Dissertação de Mestrado, apresentada à Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto/USP – Área de concentração: Biologia Comparada. -

71St Annual Meeting Society of Vertebrate Paleontology Paris Las Vegas Las Vegas, Nevada, USA November 2 – 5, 2011 SESSION CONCURRENT SESSION CONCURRENT
ISSN 1937-2809 online Journal of Supplement to the November 2011 Vertebrate Paleontology Vertebrate Society of Vertebrate Paleontology Society of Vertebrate 71st Annual Meeting Paleontology Society of Vertebrate Las Vegas Paris Nevada, USA Las Vegas, November 2 – 5, 2011 Program and Abstracts Society of Vertebrate Paleontology 71st Annual Meeting Program and Abstracts COMMITTEE MEETING ROOM POSTER SESSION/ CONCURRENT CONCURRENT SESSION EXHIBITS SESSION COMMITTEE MEETING ROOMS AUCTION EVENT REGISTRATION, CONCURRENT MERCHANDISE SESSION LOUNGE, EDUCATION & OUTREACH SPEAKER READY COMMITTEE MEETING POSTER SESSION ROOM ROOM SOCIETY OF VERTEBRATE PALEONTOLOGY ABSTRACTS OF PAPERS SEVENTY-FIRST ANNUAL MEETING PARIS LAS VEGAS HOTEL LAS VEGAS, NV, USA NOVEMBER 2–5, 2011 HOST COMMITTEE Stephen Rowland, Co-Chair; Aubrey Bonde, Co-Chair; Joshua Bonde; David Elliott; Lee Hall; Jerry Harris; Andrew Milner; Eric Roberts EXECUTIVE COMMITTEE Philip Currie, President; Blaire Van Valkenburgh, Past President; Catherine Forster, Vice President; Christopher Bell, Secretary; Ted Vlamis, Treasurer; Julia Clarke, Member at Large; Kristina Curry Rogers, Member at Large; Lars Werdelin, Member at Large SYMPOSIUM CONVENORS Roger B.J. Benson, Richard J. Butler, Nadia B. Fröbisch, Hans C.E. Larsson, Mark A. Loewen, Philip D. Mannion, Jim I. Mead, Eric M. Roberts, Scott D. Sampson, Eric D. Scott, Kathleen Springer PROGRAM COMMITTEE Jonathan Bloch, Co-Chair; Anjali Goswami, Co-Chair; Jason Anderson; Paul Barrett; Brian Beatty; Kerin Claeson; Kristina Curry Rogers; Ted Daeschler; David Evans; David Fox; Nadia B. Fröbisch; Christian Kammerer; Johannes Müller; Emily Rayfield; William Sanders; Bruce Shockey; Mary Silcox; Michelle Stocker; Rebecca Terry November 2011—PROGRAM AND ABSTRACTS 1 Members and Friends of the Society of Vertebrate Paleontology, The Host Committee cordially welcomes you to the 71st Annual Meeting of the Society of Vertebrate Paleontology in Las Vegas. -

Flatback Turtles Along the Stunning Coastline of Western Australia
PPRROOJJEECCTT RREEPPOORRTT Expedition dates: 8 – 22 November 2010 Report published: October 2011 Beach combing for conservation: monitoring flatback turtles along the stunning coastline of Western Australia. 0 BEST BEST FOR TOP BEST WILDLIFE BEST IN ENVIRONMENT TOP HOLIDAY © Biosphere Expeditions VOLUNTEERING GREEN-MINDED RESPONSIBLE www.biosphereVOLUNTEERING-expeditions.orgSUSTAINABLE AWARD FOR NATURE ORGANISATION TRAVELLERS HOLIDAY HOLIDAY TRAVEL Germany Germany UK UK UK UK USA EXPEDITION REPORT Beach combing for conservation: monitoring flatback turtles along the stunning coastline of Western Australia. Expedition dates: 8 - 22 November 2010 Report published: October 2011 Authors: Glenn McFarlane Conservation Volunteers Australia Matthias Hammer (editor) Biosphere Expeditions This report is an adaptation of “Report of 2010 nesting activity for the flatback turtle (Natator depressus) at Eco Beach Wilderness Retreat, Western Australia” by Glenn McFarlane, ISBN: 978-0-9807857-3-9, © Conservation Volunteers. Glenn McFarlane’s report is reproduced with minor adaptations in the abstract and chapter 2; the remainder is Biosphere Expeditions’ work. All photographs in this report are Glenn McFarlane’s copyright unless otherwise stated. 1 © Biosphere Expeditions www.biosphere-expeditions.org Abstract The nesting population of Australian flatback (Natator depressus) sea turtles at Eco Beach, Western Australia, continues to be the focus of this annual programme, which resumes gathering valuable data on the species, dynamic changes to the nesting environment and a strong base for environmental teaching and training of all programme participants. Whilst the Eco Beach population is not as high in density as other Western Australian nesting populations at Cape Domett, Barrow Island or the Pilbara region, it remains significant for the following reasons: The 12 km nesting beach and survey area is free from human development, which can impact on nesting turtles and hatchlings. -

Universidad Nacional Del Comahue Centro Regional Universitario Bariloche
Universidad Nacional del Comahue Centro Regional Universitario Bariloche Título de la Tesis Microanatomía y osteohistología del caparazón de los Testudinata del Mesozoico y Cenozoico de Argentina: Aspectos sistemáticos y paleoecológicos implicados Trabajo de Tesis para optar al Título de Doctor en Biología Tesista: Lic. en Ciencias Biológicas Juan Marcos Jannello Director: Dr. Ignacio A. Cerda Co-director: Dr. Marcelo S. de la Fuente 2018 Tesis Doctoral UNCo J. Marcos Jannello 2018 Resumen Las inusuales estructuras óseas observadas entre los vertebrados, como el cuello largo de la jirafa o el cráneo en forma de T del tiburón martillo, han interesado a los científicos desde hace mucho tiempo. Uno de estos casos es el clado Testudinata el cual representa uno de los grupos más fascinantes y enigmáticos conocidos entre de los amniotas. Su inconfundible plan corporal, que ha persistido desde el Triásico tardío hasta la actualidad, se caracteriza por la presencia del caparazón, el cual encierra a las cinturas, tanto pectoral como pélvica, dentro de la caja torácica desarrollada. Esta estructura les ha permitido a las tortugas adaptarse con éxito a diversos ambientes (por ejemplo, terrestres, acuáticos continentales, marinos costeros e incluso marinos pelágicos). Su capacidad para habitar diferentes nichos ecológicos, su importante diversidad taxonómica y su plan corporal particular hacen de los Testudinata un modelo de estudio muy atrayente dentro de los vertebrados. Una disciplina que ha demostrado ser una herramienta muy importante para abordar varios temas relacionados al caparazón de las tortugas, es la paleohistología. Esta disciplina se ha involucrado en temas diversos tales como el origen del caparazón, el origen del desarrollo y mantenimiento de la ornamentación, la paleoecología y la sistemática.