Prunus Salicina
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Planting and Aftercare of New Trees
Where to start? • Fruit plants that fit into to small spaces Producing Fruit for the Home – Apple … on dwarfing rootstocks • Most traditional and local garden centers do not identify specific rootstock ….”Dwarf”, “Semi Dwarf” Ron Perry • Eventual tree size within Dwarf and Semi Dwarf is large Professor Tree Spacing Nursery ID Hort. Department Rootstocks Eventual Height Between Trees Between Rows MSU M.27 or P.22 Dwarf 6 5 10 M.9 Dwarf 8 8 12 M.26 Dwarf 16 10 16 M.7 Semi Dwarf 18 14 22 MM.106 or 111 Semi Dwarf 20 16 22 Where to start? Where to start? • Fruit plants that fit into to small spaces – Cherry - Sour • Select desired fruit which will grow in your area. Tree Spacing Rootstocks • Determine how much space you have available. Varieties Eventual Height Between Trees Between Rows Northstar Mahaleb 10 8 12 • Select varieties which are easiest to grow. Montmorency Gi.5 or 6 12 10 12 Montmorency Mahaleb 12 10 14 – Disease or insect resistant varieties to reduce pest Montmorency Mazzard 14 12 16 pressures. Balaton Mahaleb 14 12 16 – Cherry - Sweet – Assess soil / site conditions Tree Spacing • Full sun VS shade or partial Nursery ID • Soil internal drainage Rootstocks Eventual Height Between Trees Between Rows • Weed competition (lawns are too competitive) Gi.5 Dwarf 12 12 16 Gi.6 Dwarf 14 14 16 Mahaleb Semi Dwarf 20 14 16 Mazzard Semi Dwarf 24 16 20 Average Annual Minimum Temperatures Where to start? (USDA Plant Hardiness Zone Map) Most MI fruit sites Zone 5 (-20oF to -10oF) to 6 (-10oF to 0oF) • Fruit plants that fit into to small spaces – Peach, Nectarine, Apricot and Plums – Can generally plant at a spacing of 10 ft X 15 ft* • * If trained to open center or vase shape • Closer spacing, needs to be trained in Chistmas Tree form (Vertical Axe). -

(Prunus Spp) Using Random Amplified Microsatellite Polymorphism Markers
Assessment of genetic diversity and relationships among wild and cultivated Tunisian plums (Prunus spp) using random amplified microsatellite polymorphism markers H. Ben Tamarzizt, S. Ben Mustapha, G. Baraket, D. Abdallah and A. Salhi-Hannachi Laboratory of Molecular Genetics, Immunology & Biotechnology, Faculty of Sciences of Tunis, University of Tunis El Manar, El Manar, Tunis, Tunisia Corresponding author: A. Salhi-Hannachi E-mail: [email protected] Genet. Mol. Res. 14 (1): 1942-1956 (2015) Received January 8, 2014 Accepted July 8, 2014 Published March 20, 2015 DOI http://dx.doi.org/10.4238/2015.March.20.4 ABSTRACT. The usefulness of random amplified microsatellite polymorphism markers to study the genetic diversity and relationships among cultivars belonging to Prunus salicina and P. domestica and their wild relatives (P. insititia and P. spinosa) was investigated. A total of 226 of 234 bands were polymorphic (96.58%). The 226 random amplified microsatellite polymorphism markers were screened using 15 random amplified polymorphic DNA and inter-simple sequence repeat primers combinations for 54 Tunisian plum accessions. The percentage of polymorphic bands (96.58%), the resolving power of primers values (135.70), and the polymorphic information content demonstrated the efficiency of the primers used in this study. The genetic distances between accessions ranged from 0.18 to 0.79 with a mean of 0.24, suggesting a high level of genetic diversity at the intra- and interspecific levels. The unweighted pair group with arithmetic mean dendrogram Genetics and Molecular Research 14 (1): 1942-1956 (2015) ©FUNPEC-RP www.funpecrp.com.br Genetic diversity of Tunisian plums using RAMPO markers 1943 and principal component analysis discriminated cultivars efficiently and illustrated relationships and divergence between spontaneous, locally cultivated, and introduced plum types. -

Plums on the Prairies by Rick Sawatzky
Plums on the Prairies by Rick Sawatzky Information from Literature Much has been published about pollination, pollinators, pollinizers, fertilization and fruit set in text books and periodicals. The definitions are not difficult. Pollination is the movement of pollen among compatible flowering plants (cross-pollination) or from anthers to stigmas on the same plant or different plants of the same clone (self-pollination). Many plants will self-pollinate but set very few fruit; some authors consider them self- pollinating but they are definitely not self-fruitful. Self-fruitful plants (and clones) set a crop of fruit after self-pollination; some of these plants bear fruit with no seeds (parthenocarpy); others develop seeds with embryos that are genetically identical to the parent plant (apomixis); and others produce haploid seeds that develop from an unfertilized egg cell. (When haploid seeds germinate they are very weak seedlings with only half the chromosomes of normal seedlings.) Regarding temperate zone tree fruits, self-pollination and fruit set does not mean self-fertility and the development of normal seeds. Many temperate zone small fruit species (e.g. strawberries and raspberries) are self-fertile and develop maximum yields of fruit with normal seeds as the result of self-pollination by insects. Pollinators, usually insects, are vectors of pollen movement. Pollinizers are plants which provide the appropriate pollen for other plants. Fertilization is the process in which gametes from the pollen unite with egg cells in the ovary of the flower. Normal seeds are usually the result of this process. Also, the principles are easily understood. Poor fertilization in plums and other Prunus species results in a poor fruit set. -

Investigations of the Plum Pox Virus in Chile in the Past 20 Years
REVIEW Investigations of the Plum pox virus in Chile in the past 20 years Guido Herrera1 Sharka disease, which is caused by Plum pox virus (PPV), is one of the most serious diseases affecting stone fruit trees around the world. Identified in Bulgaria in 1931, it was restricted to the European continent until 1992 when the virus was identified in Chile. It was subsequently verified in the USA, Canada, and Argentina. After 20 years since first detecting PPV in Chile, it seems clear that the disease cannot be eradicated in spite of various measures. Considering the seriousness of this problem for the domestic industry, a series of studies have been conducted to determine the distribution and degree of transmission of the disease, its biological and molecular characterization and epidemiological aspects, etc. The available information has allowed national phytosanitary control agencies to take steps to decrease the effects of the virus. However, there is a lack of data with respect to epidemiological factors for a more accurate understanding of the performance of the virus under Chilean conditions. Key words: Sharka disease, virus, stone fruit. INTRODUCTION more precise diagnosis techniques like Polymerase Chain Reaction (PCR) (Wetzel et al., 1991; Hadidi and Levy, The first symptoms of Sharka or Pox were observed by 1994), resulting in greater knowledge about the range of farmers in southwest Bulgaria after the First World War hosts and viral strains. As well, biotechnological methods and the first scientist to describe the viral nature of the associated with genetic transformation generated plant disease was Dimitar Atanasov in 1933 (Dzhuvinov et al., varieties with characteristics of immunity to the virus 2007), calling it Sharka disease or Plum pox virus (PPV). -

Efficacy and Safety of Prunus Mume and Choline in Patients with Abnormal Level of Liver Function Test
International Journal of Clinical Trials Aslam MN et al. Int J Clin Trials. 2020 Aug;7(3):170-175 http://www.ijclinicaltrials.com pISSN 2349-3240 | eISSN 2349-3259 DOI: http://dx.doi.org/10.18203/2349-3259.ijct20203103 Original Research Article Efficacy and safety of Prunus mume and choline in patients with abnormal level of liver function test Muhammad N. Aslam1, Anwar Ali2, Suresh Kumar3* 1Department of Gastroenterology, Mayo Hospital, Lahore, Pakistan 2Department of Medicine, Saidu Group of Teaching Hospital, Khyber Pakhtunkhwa, Pakistan 3Department of Medicine, Rafa-e-Aam Hospital, Sindh, Pakistan Received: 18 March 2020 Revised: 27 April 2020 Accepted: 30 April 2020 *Correspondence: Dr. Suresh Kumar, E-mail: [email protected] Copyright: © the author(s), publisher and licensee Medip Academy. This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Background: The objectives of the study were to determine the efficacy and safety of Prunus mume and choline in patients with abnormal liver function test. Methods: This open labelled, multi-centered observational study was done for a from May 2019 to December 2019. Patients of either gender, above 18 years of age having elevated levels of aspartate aminotransferase, alanine transaminase or gamma-glutamyltransferase were included in the study after taking informed consent. One to two tablets of revolic per day were given preferably in the morning with breakfast or as per instructions of the physician. Patient follow-ups were done at 2nd and 4th week after treatment. -

Prunus Mume Japanese Apricot1 Edward F
Fact Sheet ST-512 October 1994 Prunus mume Japanese Apricot1 Edward F. Gilman and Dennis G. Watson2 INTRODUCTION Japanese Flowering Apricot may be the longest- lived of the flowering fruit trees eventually forming a gnarled, picturesque, 20-foot-tall tree (Fig. 1). Appearing during the winter on bare branches are the multitude of small, fragrant, pink flowers which add to the uniqueness of the tree’s character. The small yellow fruits which follow the blooms are inedible but attractive. GENERAL INFORMATION Scientific name: Prunus mume Pronunciation: PROO-nus MEW-may Common name(s): Japanese Apricot Family: Rosaceae USDA hardiness zones: 6 through 8 (Fig. 2) Origin: not native to North America Uses: recommended for buffer strips around parking lots or for median strip plantings in the highway; near a deck or patio; specimen; no proven urban tolerance Availability: somewhat available, may have to go out of the region to find the tree DESCRIPTION Figure 1. Young Japanese Apricot. Height: 12 to 20 feet Spread: 15 to 20 feet Foliage Crown uniformity: irregular outline or silhouette Crown shape: round; vase shape Leaf arrangement: alternate (Fig. 3) Crown density: moderate Leaf type: simple Growth rate: medium Leaf margin: serrate Texture: fine Leaf shape: ovate 1. This document is adapted from Fact Sheet ST-512, a series of the Environmental Horticulture Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. Publication date: October 1994. 2. Edward F. Gilman, associate professor, Environmental Horticulture Department; Dennis G. Watson, associate professor, Agricultural Engineering Department, Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida, Gainesville FL 32611. -

Genome-Wide SNP Identification in Prunus Rootstocks Germplasm
www.nature.com/scientificreports OPEN Genome-wide SNP identifcation in Prunus rootstocks germplasm collections using Genotyping-by- Sequencing: phylogenetic analysis, distribution of SNPs and prediction of their efect on gene function Verónica Guajardo1, Simón Solís1, Rubén Almada1, Christopher Saski2, Ksenija Gasic2 & María Ángeles Moreno3* Genotyping-by-Sequencing (GBS) was applied in a set of 53 diploid Prunus rootstocks and fve scion cultivars from three subgenera (Amygdalus, Prunus and Cerasus) for genome-wide SNP identifcation and to assess genetic diversity of both Chilean and Spanish germplasm collections. A group of 45,382 high quality SNPs (MAF >0.05; missing data <5%) were selected for analysis of this group of 58 accessions. These SNPs were distributed in genic and intergenic regions in the eight pseudomolecules of the peach genome (Peach v2.0), with an average of 53% located in exonic regions. The genetic diversity detected among the studied accessions divided them in three groups, which are in agreement with their current taxonomic classifcation. SNPs were classifed based on their putative efect on annotated genes and KOG analysis was carried out to provide a deeper understanding of the function of 119 genes afected by high-impact SNPs. Results demonstrate the high utility for Prunus rootstocks identifcation and studies of diversity in Prunus species. Also, given the high number of SNPs identifed in exonic regions, this strategy represents an important tool for fnding candidate genes underlying traits of interest and potential functional markers for use in marker-assisted selection. Prunus is a genus belonging to the subfamily Prunoideae of the family Rosaceae1. Several species of this large genus, known as stone fruits, are among the most important for the world fruit industry, providing edible and tasty fruits highly appreciated by consumers (e.g., peaches, plums, cherries, apricots and almonds). -

Plant Gems from China©
1 Plant Gems from China© Donghui Peng1, Longqing Chen2 and Mengmeng Gu3 1College of Landscape Architecture and Horticulture, Fujian Agriculture and Forestry University, Fuzhou, Fujian Province 350002, PRC 2College of Forestry and Horticulture, Huazhong Agriculture University, Wuhan, Hubei Province 430070, PRC 3Department of Horticultural Sciences, Texas A&M AgriLife Extension Service, College Station, TX 77843, USA Email: [email protected] INTRODUCTION A lot of plants native in China thrive in landscapes across the U.S. Chinese plant germplasm has been continuously introduced to the U.S., and used in breeding and selection. So many new cultivars with Chinese genetics have been introduced in the landscape plant market. The Chinese love plants and particularly enjoy ten “traditionally famous flowers”: lotus (Nelumbo nucifera), sweet olive (Osmanthus frangrans), peony (Paeonia suffruticosa), azalea (Azalea spp.), chrysanthemum (Chrysanthemum spp.), Mei flower (Prunus mume), daffodil (Narcissus spp.), rose (Rosa spp.), camellia (Camellia spp.) and cymbidium (Cymbidium spp.). Public and university breeders have focused on these taxa. In addition, many species and cultivars commonly grown in China may be of interest to growers and landscape professionals in the U.S, which this manuscript will be focused on. PLANT SPECIES AND CULTIVARS Sweet olive (Osmanthus fragrans). There are mainly four types of sweet olives, Auranticus Group, Luteus Group, Albus Group, orange and Semperflorens Group. Ever-blooming sweet 1 2 olives have peak blooming in the fall like the others, and continue for about six months although not as profusely. Recently there are three variegated cultivars: ‘Yinbian Caiye’ with white leaf margins mature leaves and red/white/green on new growth, ‘Yintian Cai’ with red-margined maroon leaves maturing to white-margined green leaves, and ‘Pearl Color’ with pink new growth. -

Downloading Or Purchasing Online At
Commercialisation of Mume in Australia April 2015 RIRDC Publication No. 15/044 Commercialisation of mume in Australia by Bruce Topp, Dougal Russell, Grant Bignell, Janelle Dahler, Janet Giles and Judy Noller March 2015 RIRDC Publication No 15/044 RIRDC Project No PRJ-000857 © 2015 Rural Industries Research and Development Corporation. All rights reserved. ISBN 978-1-74254-788-6 ISSN 1440-6845 Commercialisation of Mume in Australia Publication No. 15/044 Project No. PRJ-000857 The information contained in this publication is intended for general use to assist public knowledge and discussion and to help improve the development of sustainable regions. You must not rely on any information contained in this publication without taking specialist advice relevant to your particular circumstances. While reasonable care has been taken in preparing this publication to ensure that information is true and correct, the Commonwealth of Australia gives no assurance as to the accuracy of any information in this publication. The Commonwealth of Australia, the Rural Industries Research and Development Corporation (RIRDC), the authors or contributors expressly disclaim, to the maximum extent permitted by law, all responsibility and liability to any person, arising directly or indirectly from any act or omission, or for any consequences of any such act or omission, made in reliance on the contents of this publication, whether or not caused by any negligence on the part of the Commonwealth of Australia, RIRDC, the authors or contributors. The Commonwealth of Australia does not necessarily endorse the views in this publication. This publication is copyright. Apart from any use as permitted under the Copyright Act 1968, all other rights are reserved. -

Plum Prunus Domestica L.; Prunus Salicina Lindl
Plum Prunus domestica L.; Prunus salicina Lindl. Rosaceae Species description There are two primary species of cultivated plums, the European plum (Prunus domestica) and Japanese plum (Prunus salicina). Plum trees are small to medium-sized deciduous trees with rounded crowns. Plum leaves are oval to elliptic in shape depending on the species, with serrated margins. The flower buds form with leaf buds in early spring. Large, white flowers appear in April. European plums have 1-2 flowers per bud, and Japanese plums generally have multiple flowers per bud. Plum fruit, a drupe fruit with a single seed, matures from July to November, depending on the cultivar. The fruit skin is covered in a smooth, waxy layer, and the skin color may be green, yellow, orange, pink, red, purple, or blue. Interior flesh color varies from green to yellow to red. European plums are usually small, oval in shape and variable in color; these include varieties such as the prune-plums Stanley and Italian, Yellow egg, Reine Claude, and the Gage plums. Japanese plums are large, round to heart- shaped and have firm flesh; these are the most common fresh eating plums in the U.S. Natural and cultural history European plums are native to western Asia in the Caucasus Mountains. Japanese plums are thought to have originated in China on the Yangtze River; widespread cultivation for the last 2,500 years makes it difficult to determine its natural range. European plum pits have been found in archaeological remains in Switzerland. Plums are mentioned in Greek writings and were common in European gardens since at least the 1st century CE. -

A Systematic Review of Ume Health Benefits
St. Catherine University SOPHIA Master of Arts in Holistic Health Studies Research Papers Holistic Health Studies 5-2016 A Systematic Review of Ume Health Benefits Mina Christensen St. Catherine University Follow this and additional works at: https://sophia.stkate.edu/ma_hhs Part of the Alternative and Complementary Medicine Commons Recommended Citation Christensen, Mina. (2016). A Systematic Review of Ume Health Benefits. Retrieved from Sophia, the St. Catherine University repository website: https://sophia.stkate.edu/ma_hhs/4 This Thesis is brought to you for free and open access by the Holistic Health Studies at SOPHIA. It has been accepted for inclusion in Master of Arts in Holistic Health Studies Research Papers by an authorized administrator of SOPHIA. For more information, please contact [email protected]. Running head: A SYTEMATIC REVIEW OF UME HEALTH BENEFITS 1 A Systematic Review of Ume Health Benefits Mina Christensen St. Catherine University May 18, 2016 A SYTEMATIC REVIEW OF UME HEALTH BENEFITS ii Abstract Ume is a Japanese apricot, and Bainiku Ekisu (BE) is concentrated ume juice. Ume has been used as a food and folk remedy for thousands of years in East Asian countries, and BE was created in Japan centuries ago and has been used as a remedy since. In the Western world, the nutritional and medicinal benefits of ume and BE are not popularly known, despite many experimental studies showing their numerous health benefits. There are no known systematic reviews of the health benefits of ume and BE. Therefore, this systematic review describes the last 20 years of quality empirical studies that investigate ume and BE health benefits. -
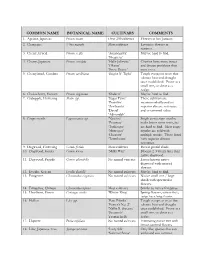
Small Maturing Tree List
COMMON NAME BOTANICAL NAME CULTIVARS COMMENTS 1. Apricot, Japanese Prunus mume Over 250 cultivars Flowers in late January. 2. Chastetree Vitex negundo Most cultivars Lavender flowers in summer. 3. Cherry, hybrid Prunus x spp. ‘Amanogawa’ May be hard to find. ‘Shogetsu’ 4. Cherry, Japanese Prunus serrulata ‘Hally Jolivette’ Cherries have more insect ‘Okame’ and disease problems than ‘Snow Goose’ most trees. 5. Cherrylaurel, Carolina Prunus caroliniana ‘Bright N’ Tight’ Tough evergreen trees that tolerate heat and drought once established. Prune as a small tree, or shear as a hedge. 6. Chokecherry, Eastern Prunus virginiana ‘Shubert’ May be hard to find. 7. Crabapple, Flowering Malus spp. ‘Sugar Tyme’ These cultivars are ‘Prairifire’ recommended based on ‘floribunda’ superior disease resistance ‘David’ and ornamental value. ‘Adirondak’ 8. Crape myrtle Lagerstroemia spp. ‘Natchez’ Single stem crape myrtles ‘Potomac’ make better street trees, but ‘Tuskeegee’ are hard to find. Most crape ‘Muscogee’ myrtles are sold with ‘Choctaw’ multiple trunks. Those listed ‘Townhouse’ have superior disease resistance. 9. Dogwood, Flowering Cornus florida Most cultivars Best in partial shade. 10. Dogwood, Kousa Cornus kousa ‘Milky Way’ Blooms 2-3 weeks later than native dogwood. 11. Dogwood, Pagoda Cornus alternifolia No named varieties Lesser known native dogwood with unusual flowers. 12. Evodia, Korean Evodia daniellii No named cultivars May be hard to find. 13. Fringetree Chionanthus virginicus No named cultivars Native small tree / large shrub with spectacular flowers. 14. Fringetree, Chinese Chionanthus refusus Most cultivars Similar to native fringetree. 15. Hawthorn, Green Crataegus viridis ‘Winter King’ Spring flowers, winter fruit; twigs have long thorns. 16. Hollies Llex spp.