Medicago Sativa L.) Populations Using Morphological Traits and Rapd Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Accepted Manuscript
Accepted Manuscript Analysis of the 2012 Ahar-Varzeghan (Iran) seismic sequence: Insights from statistical and stress transfer modeling Pouye Yazdi, Miguel Angel Santoyo, Jorge M. Gaspar-Escribano PII: S0921-8181(17)30368-5 DOI: https://doi.org/10.1016/j.gloplacha.2017.12.007 Reference: GLOBAL 2691 To appear in: Global and Planetary Change Received date: 16 July 2017 Revised date: 17 October 2017 Accepted date: 6 December 2017 Please cite this article as: Pouye Yazdi, Miguel Angel Santoyo, Jorge M. Gaspar-Escribano , Analysis of the 2012 Ahar-Varzeghan (Iran) seismic sequence: Insights from statistical and stress transfer modeling. The address for the corresponding author was captured as affiliation for all authors. Please check if appropriate. Global(2017), https://doi.org/ 10.1016/j.gloplacha.2017.12.007 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT Analysis of the 2012 Ahar-Varzeghan (Iran) Seismic Sequence: Insights from Statistical and Stress Transfer Modeling Pouye Yazdi (1); Miguel Angel Santoyo (2); Jorge M. Gaspar-Escribano (3) (1) Universidad Politécnica de Madrid (UPM), ETSI Topografía, Geodesia y Cartografía, C/Mercator 2, Campus Sur 28031 Madrid, Spain [email protected] (2) Universidad Nacional Autónoma de Mexico (UNAM), Institute of Geophysics in Campus Morelia 58190 Morelia, México [email protected] (3) Universidad Politécnica de Madrid (UPM), ETSI Topografía, Geodesia y Cartografía, C/Mercator 2, Campus Sur 28031 Madrid, Spain [email protected] Abstract The 2012 Ahar-Varzeghan (Northwestern Iran) earthquake doublet and its following seismic sequence are analyzed in this paper. -

(Mercet, 1924) (Hymenoptera: Eulophidae) in the Middle East
J. Crop Prot. 2016, 5 (2): 307-311______________________________________________________ doi: 10.18869/modares.jcp.5.2.307 Short Paper First record of Hemiptarsenus autonomus (Mercet, 1924) (Hymenoptera: Eulophidae) in the Middle East Amir-Reza Piruznia1, Hossein Lotfalizadeh2* and Mohammad-Reza Zargaran3 1. Department of Plant Protection, Islamic Azad University, Tabriz Branch, Tabriz, Iran. 2. Department of Plant Protection, East-Azarbaijan Agricultural and Natural Resources Research Center, AREEO, Tabriz, Iran. 3. Department of Forestry, Natural Resource Faculty, University of Urmia, Urmia, Iran. Abstract: Hemiptarsenus autonomus (Mercet, 1924) (Hymenoptera: Eulophidae, Eulophinae) was found for the first time outside of Europe. Studied specimen was collected by a Malaise trap in the north west of Iran, East-Azarbaijan province, Khajeh (46°38'E & 38°09'N). Current record of Hemiptarsenus species of Iran adds up to seven species. These species and their geographical distribution in Iran are listed. Keywords: Chalcidoidea, new distribution, record, Iran, fauna Introduction12 Materials and Methods Eulophidae (Hymenoptera: Chalcidoidea) of Iran Samplings were made in using the Malaise trap has been listed by Hesami et al. (2010) and Talebi in East-Azarbaijan province, Khajeh, Iran during et al. (2011). They listed 122 eulophid species summer of 2015. All the materials were from different parts of Iran including three species subsequently transferred to the laboratory at of the genus Hemiptarsenus Westwood, 1833 Department of Plant Protection, East-Azarbaijan (Hesami et al., 2010; Talebi et al., 2011). Research Center for Agriculture and Natural Recently Lotfalizadeh et al. (2015) reported Resources, Tabriz. External morphology was Hemiptarsenus waterhousii Westwood, 1833 as a illustrated using an Olympus™ SZH, equipped parasitoid of alfalfa leaf miners in the northwest with a Canon™ A720 digital camera. -

Analyses of Maragheh Tile School One of the Most Effective Factors in Cultural Heritage Formation of Maragheh Architecture During Historical Periods
WSEAS International Conference on CULTURAL HERITAGE AND TOURISM (CUHT'08), Heraklion, Crete Island, Greece, July 22-24, 2008 Analyses of Maragheh Tile School One of the Most Effective Factors in Cultural Heritage Formation of Maragheh Architecture during Historical Periods 1AHADOLLAH A'ZAMI, 2SEYYED HOSEIN YASREBI 1Department of Architecture, 2Department of Civil Engineering 1Islamic Azad University-Heris Branch, 2Islamic Azad University-Tabriz Branch Department of Architecture, Islamic Azad University-Heris Branch, Heris City, East Azarbaijan Province IRAN Abstract: - Regarding all remained tiles of tile decoration art, it could be said that Maragheh has been one of the most important center of this art during Ilkhanid times in Iran. There have been some well known and main centers of tile works in this period such as Kashan, Takht-e-Soleiman, Isfahan, Kerman and Soltaniyeh. But in decoration field with brick and tile, around sixteen kinds of tile scrape with extraordinary diversity are noticed in Maragheh observatory. As far as color is considered; blue, cerulean, white and black colors are used in Soltaniyeh and we have mostly turquoise and brown color in Maragheh. Investigation Maragheh observatory two very valuable tiles are found: 1. square sun-dries tiles with embossed intersecting spirals which have special colors and radiance in front of sun light and give special beauty to building. 2. Big sun dried tiles with inscriptions which are voted high among the tile of the period for their size, inscriptions and the kind of writing on them. As for the first kind tiles, it should be said that, not a one tile of this kind is found in other places of Maragheh or even other places other than Maragheh during Ilkhanid times. -

And “Climate”. Qarah Dagh in Khorasan Ostan on the East of Iran 1
IRAN STATISTICAL YEARBOOK 1397 1. LAND AND CLIMATE Introduction T he statistical information that appeared in this of Tehran and south of Mazandaran and Gilan chapter includes “geographical characteristics and Ostans, Ala Dagh, Binalud, Hezar Masjed and administrative divisions” ,and “climate”. Qarah Dagh in Khorasan Ostan on the east of Iran 1. Geographical characteristics and aministrative and joins Hindu Kush mountains in Afghanistan. divisions The mountain ranges in the west, which have Iran comprises a land area of over 1.6 million extended from Ararat mountain to the north west square kilometers. It lies down on the southern half and the south east of the country, cover Sari Dash, of the northern temperate zone, between latitudes Chehel Cheshmeh, Panjeh Ali, Alvand, Bakhtiyari 25º 04' and 39º 46' north, and longitudes 44º 02' and mountains, Pish Kuh, Posht Kuh, Oshtoran Kuh and 63º 19' east. The land’s average height is over 1200 Zard Kuh which totally form Zagros ranges. The meters above seas level. The lowest place, located highest peak of this range is “Dena” with a 4409 m in Chaleh-ye-Loot, is only 56 meters high, while the height. highest point, Damavand peak in Alborz The southern mountain range stretches from Mountains, rises as high as 5610 meters. The land Khouzestan Ostan to Sistan & Baluchestan Ostan height at the southern coastal strip of the Caspian and joins Soleyman Mountains in Pakistan. The Sea is 28 meters lower than the open seas. mountain range includes Sepidar, Meymand, Iran is bounded by Turkmenistan, the Caspian Sea, Bashagard and Bam Posht Mountains. -

Mapping and Assessing the Precipitation and Temperature Changes in Arasbaran Forest Ecosystem Under Climate Change, NW of Iran
Journal of Environmental Sciences Studies (JESS) Volume 5 , Number 2, Summer(2020), 2681-2692 Journal of Environmental Sciences Studies Journal home page: www.jess.ir Mapping and Assessing the Precipitation and Temperature Changes in Arasbaran Forest Ecosystem under Climate Change, NW of Iran Roya Abedia*, Ladan Kazemi Radb, *a . Assistant professor, Department of forestry, Ahar faculty of agriculture and natural resources, University of Tabriz b . Environmental Research Institute, Academic Center for Education, Culture & Research, Iran *Email Address : [email protected] Received: (April - 7-2020) Accepted: (May - 5 -2020) Abstract This study was conducted to perform the most parameters of weather data for current and future climate in the Arasbaran forest area including precipitation and temperature. The base climate data contained daily precipitation, minimum temperatures, and maximum temperatures parameters of four weather stations. The LARS-WG was used to simulation based on the historical climate data from 2000 to 2017 and data generating for future climate forecasting was evaluate for 2018-2030. Then, the model was evaluated after assessing the model ability in each station for all four stations by comparing the monthly means and variances of observed and generated data in all stations. From the results, the study found that the precipitation level would increase in the study area. In the case of minimum and maximum temperatures, the minimum temperature would decrease 0.2-0.3 °C in Jolfa station and will increase 0.1-0.2 °C in Tabriz and 4.5-4.6 °C in Kaleybar stations. In addition, the maximum temperature would increase 0.1-0.2 °C at both in the study area. -
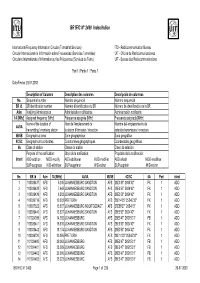
BR IFIC N° 2499 Index/Indice
BR IFIC N° 2499 Index/Indice International Frequency Information Circular (Terrestrial Services) ITU - Radiocommunication Bureau Circular Internacional de Información sobre Frecuencias (Servicios Terrenales) UIT - Oficina de Radiocomunicaciones Circulaire Internationale d'Information sur les Fréquences (Services de Terre) UIT - Bureau des Radiocommunications Part 1 / Partie 1 / Parte 1 Date/Fecha: 29.07.2003 Description of Columns Description des colonnes Descripción de columnas No. Sequential number Numéro séquenciel Número sequencial BR Id. BR identification number Numéro d'identification du BR Número de identificación de la BR Adm Notifying Administration Administration notificatrice Administración notificante 1A [MHz] Assigned frequency [MHz] Fréquence assignée [MHz] Frecuencia asignada [MHz] Name of the location of Nom de l'emplacement de Nombre del emplazamiento de 4A/5A transmitting / receiving station la station d'émission / réception estación transmisora / receptora 4B/5B Geographical area Zone géographique Zona geográfica 4C/5C Geographical coordinates Coordonnées géographiques Coordenadas geográficas 6A Class of station Classe de station Clase de estación Purpose of the notification: Objet de la notification: Propósito de la notificación: Intent ADD-addition MOD-modify ADD-additioner MOD-modifier ADD-añadir MOD-modificar SUP-suppress W/D-withdraw SUP-supprimer W/D-retirer SUP-suprimir W/D-retirar No. BR Id Adm 1A [MHz] 4A/5A 4B/5B 4C/5C 6A Part Intent 1 100039437 AFS 5.245 JOHANNESBURG SANDTON AFS 28E3'53" 26S4'42" FX 1 ADD -

See the Document
IN THE NAME OF GOD IRAN NAMA RAILWAY TOURISM GUIDE OF IRAN List of Content Preamble ....................................................................... 6 History ............................................................................. 7 Tehran Station ................................................................ 8 Tehran - Mashhad Route .............................................. 12 IRAN NRAILWAYAMA TOURISM GUIDE OF IRAN Tehran - Jolfa Route ..................................................... 32 Collection and Edition: Public Relations (RAI) Tourism Content Collection: Abdollah Abbaszadeh Design and Graphics: Reza Hozzar Moghaddam Photos: Siamak Iman Pour, Benyamin Tehran - Bandarabbas Route 48 Khodadadi, Hatef Homaei, Saeed Mahmoodi Aznaveh, javad Najaf ...................................... Alizadeh, Caspian Makak, Ocean Zakarian, Davood Vakilzadeh, Arash Simaei, Abbas Jafari, Mohammadreza Baharnaz, Homayoun Amir yeganeh, Kianush Jafari Producer: Public Relations (RAI) Tehran - Goragn Route 64 Translation: Seyed Ebrahim Fazli Zenooz - ................................................ International Affairs Bureau (RAI) Address: Public Relations, Central Building of Railways, Africa Blvd., Argentina Sq., Tehran- Iran. www.rai.ir Tehran - Shiraz Route................................................... 80 First Edition January 2016 All rights reserved. Tehran - Khorramshahr Route .................................... 96 Tehran - Kerman Route .............................................114 Islamic Republic of Iran The Railways -

Article a Catalog of Iranian Prostigmatic Mites of Superfamilies
Persian Journal of Acarology, Vol. 2, No. 3, pp. 389–474. Article A catalog of Iranian prostigmatic mites of superfamilies Raphignathoidea & Tetranychoidea (Acari) Gholamreza Beyzavi1*, Edward A. Ueckermann2 & 3, Farid Faraji4 & Hadi Ostovan1 1 Department of Entomology, Science and Research Branch, Islamic Azad University, Fars, Iran; E-mail: [email protected] 2 ARC-Plant Protection Research Institute, Private bag X123, Queenswood, Pretoria, 0121, South Africa; E-mail: [email protected] 3 School of Environmental Sciences and Development, Zoology, North-West University- Potchefstroom Campus, Potchefstroom, 2520, South Africa 4 MITOX Consultants, P. O. Box 92260, 1090 AG Amsterdam, The Netherlands * Corresponding author Abstract This catalog comprises 56 genera and 266 species of mite names of superfamilies Raphignathoidea and Tetranychoidea recorded from Iran at the end of January, 2013. Data on the mite distributions and habitats based on the published information are included. Remarks about the incorrect reports and nomen nudum species are also presented. Key words: Checklist, mite, habitat, distribution, Iran. Introduction Apparently the first checklist about mites of Iran was that of Farahbakhsh in 1961. Subsequently the following lists were published: “The 20 years researches of Acarology in Iran, List of agricultural pests and their natural enemies in Iran, A catalog of mites and ticks (Acari) of Iran and Injurious mites of agricultural crops in Iran” are four main works (Sepasgosarian 1977; Modarres Awal 1997; Kamali et al. 2001; Khanjani & Haddad Irani-Nejad 2006). Prostigmatic mites consist of parasitic, plant feeding and beneficial predatory species and is the major group of Acari in the world. Untill 2011, 26205 species were described in this suborder, of which 4728 species belong to the cohort Raphignathina and tetranychoid and raphignathoid mites include 2211 and 877 species respectively (Zhang et al. -

Life Science Journal 2015;12(7)
Life Science Journal 2015;12(7) http://www.lifesciencesite.com Feasibility Study of Tourism Development Impacts on Stable Regional Development Dimensions: The Case Study Rahim Abdollah Fam1 and Parviz Kesavarz2 1 Department of Social Science, Bonab Branch, Islamic Azad University, Bonab, Iran. 2 (Corresponding Author), Department of Management, Bonab Branch, Islamic Azad University, Bonab, Iran. E-mail: [email protected]. Abstract: Without a doubt one of the most important challenges the region programmers are facing in many countries are issues related to sustainability and sustainable regional development. There are several factors that could have influenced the sustainable development in the region. One of these factors could be tourism and its consequences. Comprehensive study of the impact of tourism in the region requires a regional study in the context of sustainable development. District of Kandovan Tourism with over 850 years of being in the western foothills of the Sahand Mountains in the East Azerbaijan province has an influential impact in attracting Tourism and regional & national development by diverse and pristine nature and cultural and historical authenticity. The present study reviews the concepts of sustainable tourism, sustainable development implications of tourism on the region. This method is based on analytic methods for data collection and data archival and field methods have been used. Statistical Society is Tourism and tourists as well as residents and officials in the region that Cochran method is being used to determine the number of samples. Achieved sample size is 341 people from a random distribution of the questionnaire. Results gathered represent that despite positive economic impact on some aspects of the social and environmental aspects, results are often negative that help to regional instability. -

Mayors for Peace Member Cities 2021/10/01 平和首長会議 加盟都市リスト
Mayors for Peace Member Cities 2021/10/01 平和首長会議 加盟都市リスト ● Asia 4 Bangladesh 7 China アジア バングラデシュ 中国 1 Afghanistan 9 Khulna 6 Hangzhou アフガニスタン クルナ 杭州(ハンチォウ) 1 Herat 10 Kotwalipara 7 Wuhan ヘラート コタリパラ 武漢(ウハン) 2 Kabul 11 Meherpur 8 Cyprus カブール メヘルプール キプロス 3 Nili 12 Moulvibazar 1 Aglantzia ニリ モウロビバザール アグランツィア 2 Armenia 13 Narayanganj 2 Ammochostos (Famagusta) アルメニア ナラヤンガンジ アモコストス(ファマグスタ) 1 Yerevan 14 Narsingdi 3 Kyrenia エレバン ナールシンジ キレニア 3 Azerbaijan 15 Noapara 4 Kythrea アゼルバイジャン ノアパラ キシレア 1 Agdam 16 Patuakhali 5 Morphou アグダム(県) パトゥアカリ モルフー 2 Fuzuli 17 Rajshahi 9 Georgia フュズリ(県) ラージシャヒ ジョージア 3 Gubadli 18 Rangpur 1 Kutaisi クバドリ(県) ラングプール クタイシ 4 Jabrail Region 19 Swarupkati 2 Tbilisi ジャブライル(県) サルプカティ トビリシ 5 Kalbajar 20 Sylhet 10 India カルバジャル(県) シルヘット インド 6 Khocali 21 Tangail 1 Ahmedabad ホジャリ(県) タンガイル アーメダバード 7 Khojavend 22 Tongi 2 Bhopal ホジャヴェンド(県) トンギ ボパール 8 Lachin 5 Bhutan 3 Chandernagore ラチン(県) ブータン チャンダルナゴール 9 Shusha Region 1 Thimphu 4 Chandigarh シュシャ(県) ティンプー チャンディーガル 10 Zangilan Region 6 Cambodia 5 Chennai ザンギラン(県) カンボジア チェンナイ 4 Bangladesh 1 Ba Phnom 6 Cochin バングラデシュ バプノム コーチ(コーチン) 1 Bera 2 Phnom Penh 7 Delhi ベラ プノンペン デリー 2 Chapai Nawabganj 3 Siem Reap Province 8 Imphal チャパイ・ナワブガンジ シェムリアップ州 インパール 3 Chittagong 7 China 9 Kolkata チッタゴン 中国 コルカタ 4 Comilla 1 Beijing 10 Lucknow コミラ 北京(ペイチン) ラクノウ 5 Cox's Bazar 2 Chengdu 11 Mallappuzhassery コックスバザール 成都(チォントゥ) マラパザーサリー 6 Dhaka 3 Chongqing 12 Meerut ダッカ 重慶(チョンチン) メーラト 7 Gazipur 4 Dalian 13 Mumbai (Bombay) ガジプール 大連(タァリィェン) ムンバイ(旧ボンベイ) 8 Gopalpur 5 Fuzhou 14 Nagpur ゴパルプール 福州(フゥチォウ) ナーグプル 1/108 Pages -

TÜBA-AR Sayı23.Pdf
Prof.Dr. Harald HAUPTMANN (4 Eylül 1936 - 2 Ağustos 2018) Saygıyla anıyoruz... In Memoriam... TÜBA-AR Türkiye Bilimler Akademisi Arkeoloji Dergisi Turkish Academy of Sciences Journal of Archaeology Sayı: 23 Volume: 23 2018 TÜBA Arkeoloji (TÜBA-AR) Dergisi TÜBA-AR TÜBA-AR uluslararası hakemli bir TÜRKİYE BİLİMLER AKADEMİSİ ARKEOLOJİ DERGİSİ dergi olup TÜBİTAK ULAKBİM (SBVT) ve Avrupa İnsani Bilimler Referans TÜBA-AR, Türkiye Bilimler Akademisi (TÜBA) tarafından yıllık olarak İndeksi (ERIH PLUS) veritabanlarında yayınlanan uluslararası hakemli bir dergidir. Derginin yayın politikası, kapsamı taranmaktadır. ve içeriği ile ilgili kararlar, Türkiye Bilimler Akademisi Konseyi tarafından TÜBA Journal of Archaeology belirlenen Yayın Kurulu tarafından alınır. (TÜBA-AR) TÜBA-AR is an international refereed journal and indexed in the TUBİTAK DERGİNİN KAPSAMI VE YAYIN İLKELERİ ULAKBİM (SBVT) and The European Reference Index for the Humanities TÜBA-AR dergisi ilke olarak, dönem ve coğrafi bölge sınırlaması olmadan and the Social Sciences (ERIH PLUS) arkeoloji ve arkeoloji ile bağlantılı tüm alanlarda yapılan yeni araştırma, yorum, databases. değerlendirme ve yöntemleri kapsamaktadır. Dergi arkeoloji alanında yeni yapılan çalışmalara yer vermenin yanı sıra, bir bilim akademisi yayın organı Sahibi / Owner: Türkiye Bilimler Akademisi adına olarak, arkeoloji ile bağlantılı olmak koşuluyla, sosyal bilimlerin tüm uzmanlık Prof. Dr. Ahmet Cevat ACAR alanlarına açıktır; bu alanlarda gelişen yeni yorum, yaklaşım, analizlere yer veren (Başkan / President) bir forum oluşturma işlevini de yüklenmiştir. Sorumlu Yazı İşleri Müdürü Dergi, arkeoloji ile ilgili yeni açılımları kapsamlı olarak ele almak için belirli Managing Editor Prof. Dr. Ahmet Nuri YURDUSEV bir konuya odaklanmış yazıları “dosya” şeklinde kapsamına alabilir; bu amaçla çağrılı yazarların katkısının istenmesi ya da bu bağlamda gelen istekler Yayın Basın ve Halkla İlişkiler Kurulu tarafından değerlendirir. -

An Integrated Approach of Analytical Network Process and Fuzzy Based Spatial Decision Making Systems Applied to Landslide Risk Mapping
Journal of African Earth Sciences 133 (2017) 15e24 Contents lists available at ScienceDirect Journal of African Earth Sciences journal homepage: www.elsevier.com/locate/jafrearsci An integrated approach of analytical network process and fuzzy based spatial decision making systems applied to landslide risk mapping * Hassan Abedi Gheshlaghi , Bakhtiar Feizizadeh Department of Remote Sensing and GIS, Faculty of Geography and Planning, University of Tabriz, Tabriz, P.O. Box 51666-16471, Iran article info abstract Article history: Landslides in mountainous areas render major damages to residential areas, roads, and farmlands. Hence, Received 20 February 2017 one of the basic measures to reduce the possible damage is by identifying landslide-prone areas through Received in revised form landslide mapping by different models and methods. The purpose of conducting this study is to evaluate 19 April 2017 the efficacy of a combination of two models of the analytical network process (ANP) and fuzzy logic in Accepted 4 May 2017 landslide risk mapping in the Azarshahr Chay basin in northwest Iran. After field investigations and a Available online 5 May 2017 review of research literature, factors affecting the occurrence of landslides including slope, slope aspect, altitude, lithology, land use, vegetation density, rainfall, distance to fault, distance to roads, distance to Keywords: Landslide risk mapping rivers, along with a map of the distribution of occurred landslides were prepared in GIS environment. Analytical network process Then, fuzzy logic was used for weighting sub-criteria, and the ANP was applied to weight the criteria. Fuzzy logic Next, they were integrated based on GIS spatial analysis methods and the landslide risk map was pro- Integration duced.