Gene-Environment Interaction and Maternal Arsenic Methylation Efficiency During Pregnancy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

A State of the Art of Antioxidant Properties of Curcuminoids in Neurodegenerative Diseases
International Journal of Molecular Sciences Review A State of the Art of Antioxidant Properties of Curcuminoids in Neurodegenerative Diseases Serena Silvestro † , Cinzia Sindona †, Placido Bramanti and Emanuela Mazzon * IRCCS Centro Neurolesi “Bonino-Pulejo”, Via Provinciale Palermo, Contrada Casazza, 98124 Messina, Italy; [email protected] (S.S.); [email protected] (C.S.); [email protected] (P.B.) * Correspondence: [email protected]; Tel.: +39-090-60128172 † These authors contributed equally as first author. Abstract: Neurodegenerative diseases represent a set of pathologies characterized by an irreversible and progressive, and a loss of neuronal cells in specific areas of the brain. Oxidative phosphorylation is a source of energy production by which many cells, such as the neuronal cells, meet their energy needs. Dysregulations of oxidative phosphorylation induce oxidative stress, which plays a key role in the onset of neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS). To date, for most neurodegenerative diseases, there are no resolute treatments, but only interventions capable of alleviating the symptoms or slowing the course of the disease. Therefore, effective neuroprotection strategies are needed. In recent years, natural products, such as curcuminoids, have been intensively explored and studied for their therapeutic potentials in several neurodegenerative diseases. Curcuminoids are, nutraceutical compouns, that owen several therapeutic properties such as anti-oxidant, anti-inflammatory and neuroprotective effects. In this context, the aim of this review was to provide an overview of preclinical and clinical Citation: Silvestro, S.; Sindona, C.; evidence aimed to illustrate the antioxidant effects of curcuminoids in neurodegenerative diseases. -

Role of GSH and Iron-Sulfur Glutaredoxins in Iron Metabolism—Review
molecules Review Role of GSH and Iron-Sulfur Glutaredoxins in Iron Metabolism—Review 1, 1, 1 1 Trnka Daniel y , Hossain Md Faruq y , Jordt Laura Magdalena , Gellert Manuela and Lillig Christopher Horst 2,* 1 Institute for Medical Biochemistry and Molecular Biology, University Medicine, University of Greifswald, 17475 Greifswald, Germany; [email protected] (T.D.); [email protected] (H.M.F.); [email protected] (J.L.M.); [email protected] (G.M.) 2 Christopher Horst Lillig, Institute for Medical Biochemistry and Molecular Biology, University Medicine Greifswald, Ferdinand-Sauerbruch-Straße, 17475 Greifswald, Germany * Correspondence: [email protected]; Tel.: +49-3834-865407; Fax: +49-3834-865402 These authors contributed equally to this work. y Academic Editor: Pál Perjési Received: 29 July 2020; Accepted: 22 August 2020; Published: 25 August 2020 Abstract: Glutathione (GSH) was initially identified and characterized for its redox properties and later for its contributions to detoxification reactions. Over the past decade, however, the essential contributions of glutathione to cellular iron metabolism have come more and more into focus. GSH is indispensable in mitochondrial iron-sulfur (FeS) cluster biosynthesis, primarily by co-ligating FeS clusters as a cofactor of the CGFS-type (class II) glutaredoxins (Grxs). GSH is required for the export of the yet to be defined FeS precursor from the mitochondria to the cytosol. In the cytosol, it is an essential cofactor, again of the multi-domain CGFS-type Grxs, master players in cellular iron and FeS trafficking. In this review, we summarize the recent advances and progress in this field. The most urgent open questions are discussed, such as the role of GSH in the export of FeS precursors from mitochondria, the physiological roles of the CGFS-type Grx interactions with BolA-like proteins and the cluster transfer between Grxs and recipient proteins. -

N6AMT1 / HEMK2 (1-214, His-Tag) Human Protein – AR51691PU-N
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for AR51691PU-N N6AMT1 / HEMK2 (1-214, His-tag) Human Protein Product data: Product Type: Recombinant Proteins Description: N6AMT1 / HEMK2 (1-214, His-tag) human recombinant protein, 0.1 mg Species: Human Expression Host: E. coli Tag: His-tag Predicted MW: 25.3 kDa Concentration: lot specific Purity: >85% by SDS - PAGE Buffer: Presentation State: Purified Buffer System: Liquid, In Phosphate buffered saline (pH7.4) containing 20% glycerol, 1mM DTT. Protein Description: Recombinant human N6ATM1 protein, fused to His-tag at N-terminus, was expressed in E.coli and purified by using conventional chromatography techniques. Storage: Store undiluted at 2-8°C for one week or (in aliquots) at -20°C to -80°C for longer. Avoid repeated freezing and thawing. Stability: Shelf life: one year from despatch. RefSeq: NP_037372 Locus ID: 29104 UniProt ID: Q9Y5N5 Cytogenetics: 21q21.3 Synonyms: C21orf127; HEMK2; KMT9; m.HsaHemK2P; MTQ2; N6AMT; PRED28; PrmC Summary: This gene encodes an N(6)-adenine-specific DNA methyltransferase. The encoded enzyme may be involved in the methylation of release factor I during translation termination. This enzyme is also involved in converting the arsenic metabolite monomethylarsonous acid to the less toxic dimethylarsonic acid. Alternative splicing pf this gene results in multiple transcript variants. A related pseudogene has been identified on chromosome 11. [provided by RefSeq, Jul 2014] Protein Families: Druggable Genome This product is to be used for laboratory only. -

N6AMT1 Rabbit Pab
Leader in Biomolecular Solutions for Life Science N6AMT1 Rabbit pAb Catalog No.: A7201 Basic Information Background Catalog No. This gene encodes an N(6)-adenine-specific DNA methyltransferase. The encoded A7201 enzyme may be involved in the methylation of release factor I during translation termination. This enzyme is also involved in converting the arsenic metabolite Observed MW monomethylarsonous acid to the less toxic dimethylarsonic acid. Alternative splicing pf 23KDa this gene results in multiple transcript variants. A related pseudogene has been identified on chromosome 11. Calculated MW 19kDa/22kDa Category Primary antibody Applications WB, IHC, IF Cross-Reactivity Human, Mouse, Rat Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 29104 Q9Y5N5 IHC 1:50 - 1:200 Immunogen 1:50 - 1:100 IF Recombinant fusion protein containing a sequence corresponding to amino acids 1-186 of human N6AMT1 (NP_877426.3). Synonyms N6AMT1;C21orf127;HEMK2;MTQ2;N6AMT;PRED28;m.HsaHemK2P Contact Product Information www.abclonal.com Source Isotype Purification Rabbit IgG Affinity purification Storage Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide,50% glycerol,pH7.3. Validation Data Western blot analysis of extracts of Rat kidney, using N6AMT1 antibody (A7201) at 1:1000 dilution. Secondary antibody: HRP Goat Anti-Rabbit IgG (H+L) (AS014) at 1:10000 dilution. Lysates/proteins: 25ug per lane. Blocking buffer: 3% nonfat dry milk in TBST. Detection: ECL Basic Kit (RM00020). Exposure time: 90s. Immunohistochemistry of paraffin- Immunohistochemistry of paraffin- Immunofluorescence analysis of PC-12 embedded rat brain using N6AMT1 embedded human kidney using N6AMT1 cells using N6AMT1 Rabbit pAb (A7201) at Antibody (A7201) at dilution of 1:100 (40x Antibody (A7201) at dilution of 1:100 (40x dilution of 1:100 (40x lens). -

Methyltransferase (AS3MT) and N-6 Adenine-Specific DNA Methyltransferase 1 (N6AMT1) Polymorphisms, Arsenic Metabolism, and Cancer Risk in a Chilean Population
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Environ Manuscript Author Mol Mutagen. Author Manuscript Author manuscript; available in PMC 2018 July 01. Published in final edited form as: Environ Mol Mutagen. 2017 July ; 58(6): 411–422. doi:10.1002/em.22104. Associations between arsenic (+3 oxidation state) methyltransferase (AS3MT) and N-6 adenine-specific DNA methyltransferase 1 (N6AMT1) polymorphisms, arsenic metabolism, and cancer risk in a Chilean population Rosemarie de la Rosa1, Craig Steinmaus1, Nicholas K Akers1, Lucia Conde1, Catterina Ferreccio2, David Kalman3, Kevin R Zhang1, Christine F Skibola1, Allan H Smith1, Luoping Zhang1, and Martyn T Smith1 1Superfund Research Program, School of Public Health, University of California, Berkeley, CA 2Pontificia Universidad Católica de Chile, Santiago, Chile, Advanced Center for Chronic Diseases, ACCDiS 3School of Public Health, University of Washington, Seattle, WA Abstract Inter-individual differences in arsenic metabolism have been linked to arsenic-related disease risks. Arsenic (+3) methyltransferase (AS3MT) is the primary enzyme involved in arsenic metabolism, and we previously demonstrated in vitro that N-6 adenine-specific DNA methyltransferase 1 (N6AMT1) also methylates the toxic iAs metabolite, monomethylarsonous acid (MMA), to the less toxic dimethylarsonic acid (DMA). Here, we evaluated whether AS3MT and N6AMT1 gene polymorphisms alter arsenic methylation and impact iAs-related cancer risks. We assessed AS3MT and N6AMT1 polymorphisms and urinary arsenic metabolites (%iAs, %MMA, %DMA) in 722 subjects from an arsenic-cancer case-control study in a uniquely exposed area in northern Chile. Polymorphisms were genotyped using a custom designed multiplex, ligation-dependent probe amplification (MLPA) assay for 6 AS3MT SNPs and 14 tag SNPs in the N6AMT1 gene. -

Urinary Arsenic Methylation Species, Arsenic Methylation Efficiency During Pregnancy and Birth Outcomes
Urinary Arsenic Methylation Species, Arsenic Methylation Efficiency during Pregnancy and Birth Outcomes The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:37945565 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Urinary Arsenic Methylation Species, Arsenic Methylation Efficiency during Pregnancy and Birth Outcomes Shangzhi Gao A Dissertation Submitted to the Faculty of The Harvard T.H. Chan School of Public Health in Partial Fulfillment of the Requirements for the Degree of Doctor of Science in the Department of Environmental Health Harvard University Boston, Massachusetts. May, 2018 Dissertation Advisor: Dr. David C. Christiani Shangzhi Gao Urinary Arsenic Methylation Species, Arsenic Methylation Efficiency during Pregnancy and Birth Outcomes Abstract Arsenic exposure from drinking water has been associated with symptoms during pregnancy, as well as negative birth outcomes. The individual’s capability to methylate arsenic modifies the risk of arsenic-related health outcomes. Bangladesh is facing the world’s most severe arsenic crisis, as well as a high prevalence of low birth weight. However, little is known about determinants of arsenic metabolism during pregnancy and its relationship to birth outcomes. Thus, we conducted this study in Bangladesh to identify determinants of arsenic methylation capability during pregnancy, and its relationship with birth outcomes. The study was based on a Bangladesh prospective reproductive cohort (Project Jeebon) of 1,613 pregnant women recruited between 2008 and 2011. -

Variant in the DCTN4 Gene with Wilson Disease
원 저 Journal of Genetic Medicine 2011;8:53-57 • DOI: 10.5734/JGM.2011.8.1.53 53 1) Association of a c.1084A>G (p.Thr362Ala)Variant in the DCTN4 Gene with Wilson Disease Robin Dong-Woo Lee1, 2, Jae-Jung Kim2, Joo-Hyun Kim2, Jong-Keuk Lee2 and Han-Wook Yoo2, 3 1Bel Air High School, Bel Air, MD, USA 2Asan Institute for Life Sciences, University of Ulsan College of Medicine, Seoul, Korea 3Department of Pediatrics, Asan Medical Center Children’s Hospital, University of Ulsan College of Medicine, Seoul, Korea Purpose: Wilson disease is an autosomal recessive disorder which causes excessive copper accumulation in the hepatic region. So far, ATP7B gene is the only disease-causing gene of Wilson disease known to date. However, ATP7B mutations have not been found in ~15% of the patients. This study was performed to identify any causative gene in Wilson disease patients without an ATP7B mutation in either allele. Materials and Methods: The sequence of the coding regions and exon-intron boundaries of the five ATP7B-interacting genes, ATOX1, COMMD1, GLRX, DCTN4, and ZBTB16, were analyzed in the 12 patients with Wilson disease. Results: Three nonsynonymous variants including c.1084A>G (p.Thr362Ala) in the exon 12 of the DCTN4 gene were identified in the patients examined. Among these, only p.Thr362Ala was predicted as possibly damaging protein function by in silico analysis. Examination of allele frequency of c.1084A>G (p.Thr362Ala) variant in the 176 patients with Wilson disease and in the 414 normal subjects revealed that the variant was more prevalent in the Wilson disease patients (odds ratio [OR]=3.14, 95% confidence interval=1.36-7.22, P =0.0094). -

Genetic and Non-Genetic Factors Associated with the Phenotype Of
www.nature.com/scientificreports OPEN Genetic and non‑genetic factors associated with the phenotype of exceptional longevity & normal cognition Bin Han1, Huashuai Chen2,3, Yao Yao7, Xiaomin Liu4,5, Chao Nie4,5, Junxia Min6, Yi Zeng2,7* & Michael W. Lutz8* In this study, we split 2156 individuals from the Chinese Longitudinal Healthy Longevity Survey (CLHLS) data into two groups, establishing a phenotype of exceptional longevity & normal cognition versus cognitive impairment. We conducted a genome‑wide association study (GWAS) to identify signifcant genetic variants and biological pathways that are associated with cognitive impairment and used these results to construct polygenic risk scores. We elucidated the important and robust factors, both genetic and non‑genetic, in predicting the phenotype, using several machine learning models. The GWAS identifed 28 signifcant SNPs at p‑value < 3 × 10−5 signifcance level and we pinpointed four genes, ESR1, PHB, RYR3, GRIK2, that are associated with the phenotype though immunological systems, brain function, metabolic pathways, infammation and diet in the CLHLS cohort. Using both genetic and non‑genetic factors, four machine learning models have close prediction results for the phenotype measured in Area Under the Curve: random forest (0.782), XGBoost (0.781), support vector machine with linear kernel (0.780), and ℓ2 penalized logistic regression (0.780). The top four important and congruent features in predicting the phenotype identifed by these four models are: polygenic risk score, sex, age, and education. Cognitive Impairment (CI) is defned as the loss of ability in cognitive functions, such as remembering, learning, and concentrating, which negatively impacts afected individuals’ daily activities1. -

A Potential Therapeutic Role for Angiotensin Converting Enzyme 2 in Human Pulmonary Arterial Hypertension
ERJ Express. Published on June 14, 2018 as doi: 10.1183/13993003.02638-2017 Early View Original article A potential therapeutic role for Angiotensin Converting Enzyme 2 in human pulmonary arterial hypertension Anna R. Hemnes, Anandharajan Rathinasabapathy, Eric A. Austin, Evan L. Brittain, Erica J. Carrier, Xinping Chen, Joshua P. Fessel, Candice D. Fike, Peter Fong, Niki Fortune, Robert E. Gerszten, Jennifer A. Johnson, Mark Kaplowitz, John H. Newman, Robert Piana, Meredith E. Pugh, Todd W. Rice, Ivan M. Robbins, Lisa Wheeler, Chang Yu, James E. Loyd, James West Please cite this article as: Hemnes AR, Rathinasabapathy A, Austin EA, et al. A potential therapeutic role for Angiotensin Converting Enzyme 2 in human pulmonary arterial hypertension. Eur Respir J 2018; in press (https://doi.org/10.1183/13993003.02638-2017). This manuscript has recently been accepted for publication in the European Respiratory Journal. It is published here in its accepted form prior to copyediting and typesetting by our production team. After these production processes are complete and the authors have approved the resulting proofs, the article will move to the latest issue of the ERJ online. Copyright ©ERS 2018 Copyright 2018 by the European Respiratory Society. A potential therapeutic role for Angiotensin Converting Enzyme 2 in human pulmonary arterial hypertension Anna R. Hemnes, MD*1, Anandharajan Rathinasabapathy, PhD*1, Eric A. Austin, MD, MSCI2, Evan L. Brittain, MD, MSCI3, Erica J. Carrier, PhD1, Xinping Chen, PhD1, Joshua P. Fessel, MD, PhD1, Candice D. Fike, MD2, Peter Fong, MD3, Niki Fortune1, Robert E. Gerszten, MD4, Jennifer A. Johnson, MD1, Mark Kaplowitz2, John H. -
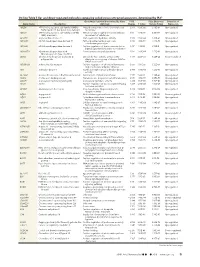
Up- and Down-Regulated Molecules Comparing Coiled Versus
On-line Table 1: Up- and down-regulated molecules comparing coiled versus untreated aneurysms, determined by IPAa Gene Main Function Determined by Gene Fold False Discovery Direction of Gene Name Description Ontology Change P Value Rate (q Value) Expression ABCB6 ATP-binding cassette, sub-family B (MDR/ Integral component of mitochondrial outer 2.541 4.28E-05 5.90E-04 Up-regulated TAP), member 6 (Langereis blood group) membrane ABCC1 ATP-binding cassette, sub-family C (CFTR/ ATPase activity, coupled to transmembrane 3.132 4.94E-10 6.17E-08 Up-regulated MRP), member 1 movement of substances ACOT11 Acyl-coa thioesterase 11 Carboxylic ester hydrolase activity 3.973 2.58E-04 2.42E-03 Up-regulated ADAM10 ADAM metallopeptidase domain 10 PMA-inducible membrane protein 3.425 1.33E-04 1.44E-03 Up-regulated ectodomain proteolysis ADAM8 ADAM metallopeptidase domain 8 Positive regulation of tumor necrosis factor 3.417 1.42E-13 6.74E-11 Up-regulated (ligand) superfamily member 11 production ADAMTS4 ADAM metallopeptidase with Proteinaceous extracellular matrix 2.134 2.49E-08 1.52E-06 Up-regulated thrombospondin type 1 motif, 4 ADH4 Alcohol dehydrogenase 4 (class II), pi Oxidoreductase activity, acting on the Ϫ2.657 4.44E-05 6.04E-04 Down-regulated polypeptide aldehyde or oxo group of donors, NAD or NADP as acceptor ADORA2B Adenosine A2b receptor Positive regulation of chronic inflammatory 2.056 1.95E-05 3.22E-04 Up-regulated response to non-antigenic stimulus AK4 Adenylate kinase 4 Nucleoside triphosphate adenylate kinase 2.074 1.96E-08 1.25E-06 Up-regulated