Bond Energies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Dependence of the Sio Bond Length on Structural Parameters in Coesite, the Silica Polymorphs, and the Clathrasils
American Mineralogist, Volume 75, pages 748-754, 1990 The dependence of the SiO bond length on structural parameters in coesite, the silica polymorphs, and the clathrasils M. B. BOISEN, JR. Department of Mathematics, Virginia Polytechnic Institute and State University, Blacksburg, Virginia 24061, U.S.A. G. V. GIBBS, R. T. DOWNS Department of Geological Sciences, Virginia Polytechnic Institute and State University, Blacksburg, Virginia 24061, U.S.A. PHILIPPE D' ARCO Laboratoire de Geologie, Ecole Normale Superieure, 75230 Paris Cedex OS, France ABSTRACT Stepwise multiple regression analyses of the apparent R(SiO) bond lengths were com- pleted for coesite and for the silica polymorphs together with the clathrasils as a function of the variables};(O), P (pressure), f,(Si), B(O), and B(Si). The analysis of 94 bond-length data recorded for coesite at a variety of pressures and temperatures indicates that all five of these variables make a significant contribution to the regression sum of squares with an R2 value of 0.84. A similar analysis of 245 R(SiO) data recorded for silica polymorphs and clathrasils indicate that only three of the variables (B(O), };(O), and P) make a signif- icant contribution to the regression with an R2 value of 0.90. The distribution of B(O) shows a wide range of values from 0.25 to 10.0 A2 with nearly 80% of the observations clustered between 0.25 and 3.0 A2 and the remaining values spread uniformly between 4.5 and 10.0 A2. A regression analysis carried out for these two populations separately indicates, for the data set with B(O) values less than three, that };(O) B(O), P, and };(Si) are all significant with an R2 value of 0.62. -

The Nafure of Silicon-Oxygen Bonds
AmericanMineralogist, Volume 65, pages 321-323, 1980 The nafure of silicon-oxygenbonds LtNus PeurrNc Linus Pauling Institute of Scienceand Medicine 2700 Sand Hill Road, Menlo Park, California 94025 Abstract Donnay and Donnay (1978)have statedthat an electron-densitydetermination carried out on low-quartz by R. F. Stewart(personal communication), which assignscharges -0.5 to the oxygen atoms and +1.0 to the silicon atoms, showsthe silicon-oxygenbond to be only 25 percentionic, and hencethat "the 50/50 descriptionofthe past fifty years,which was based on the electronegativity difference between Si and O, is incorrect." This conclusion, however, ignoresthe evidencethat each silicon-oxygenbond has about 55 percentdouble-bond char- acter,the covalenceof silicon being 6.2,with transferof 2.2 valenc,eelectrons from oxygen to silicon. Stewart'svalue of charge*1.0 on silicon then leads to 52 percentionic characterof the bonds, in excellentagreement with the value 5l percent given by the electronegativity scale. Donnay and Donnay (1978)have recently referred each of the four surrounding oxygen atoms,this ob- to an "absolute" electron-densitydetermination car- servationleads to the conclusionthat the amount of ried out on low-quartz by R. F. Stewart, and have ionic character in the silicon-oxygsa 5ingle bond is statedthat "[t showsthe Si-O bond to be 75 percent 25 percent. It is, however,not justified to make this covalent and only 25 percent ionic; the 50/50 de- assumption. scription of the past fifty years,which was basedon In the first edition of my book (1939)I pointed out the electronegativitydifference between Si and O, is that the observedSi-O bond length in silicates,given incorrect." as 1.604, is about 0.20A less than the sum of the I formulated the electronegativityscale in 1932, single-bond radii of the two atoms (p. -
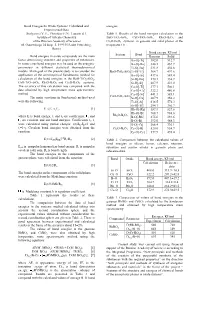
Bond Energies in Oxide Systems: Calculated and Experimental Data
Bond Energies In Oxide Systems: Calculated and energies. Experimental Data Stolyarova V. L., Plotnikov E.N., Lopatin S.I. Table 1. Results of the bond energies calculation in the Institute of Silicate Chemistry BaO-TiO 2-SiO 2, CaO-TiO 2-SiO 2, Rb 2O-B2O3 and of the Russian Academy of Sciences Cs 2O-B2O3 systems. in gaseous and solid phases at the ul. Odoevskogo 24 korp. 2, 199155 Sankt Petersburg, temperature 0 ¥ . Russia Bond energy, KJ/mol System Bond Bond energies in oxide compounds are the main Gaseous Solid factor determining structure and properties of substances. Ba-O[-Ti] 382.0 512.7 In some cases bond energies may be used as the energetic Ba-O[-Ba] 348.5 467.7 parameters in different statistical thermodynamical Ti-O[-Ba] 331.8 450.6 models. Main goal of the present study is to consider the BaO-TiO 2-SiO 2 Ti-O[-Ti] 364.7 497.6 application of the semiempirical Sanderson's method for Ba-O[-Si] 437.6 585.4 calculation of the bond energies in the BaO-TiO 2-SiO 2, Si-O[-Ba] 270.3 334.7 CaO-TiO 2-SiO 2, Rb 2O-B2O3 and Cs 2O-B2O3 systems. Si-O[-Si] 407.9 421.0 The accuracy of this calculation was compared with the Ca-O[-Ti] 377.1 500.1 data obtained by high temperature mass spectrometric Ca-O[-Ca] 322.2 486.8 method. Ca-O[-Si] 441.1 585.1 CaO-TiO -SiO The main equations in Sanderson's method used 2 2 Si-O[-Ca] 287.9 358.2 were the following. -

Molecular Orbital Theory to Predict Bond Order • to Apply Molecular Orbital Theory to the Diatomic Homonuclear Molecule from the Elements in the Second Period
Skills to Develop • To use molecular orbital theory to predict bond order • To apply Molecular Orbital Theory to the diatomic homonuclear molecule from the elements in the second period. None of the approaches we have described so far can adequately explain why some compounds are colored and others are not, why some substances with unpaired electrons are stable, and why others are effective semiconductors. These approaches also cannot describe the nature of resonance. Such limitations led to the development of a new approach to bonding in which electrons are not viewed as being localized between the nuclei of bonded atoms but are instead delocalized throughout the entire molecule. Just as with the valence bond theory, the approach we are about to discuss is based on a quantum mechanical model. Previously, we described the electrons in isolated atoms as having certain spatial distributions, called orbitals, each with a particular orbital energy. Just as the positions and energies of electrons in atoms can be described in terms of atomic orbitals (AOs), the positions and energies of electrons in molecules can be described in terms of molecular orbitals (MOs) A particular spatial distribution of electrons in a molecule that is associated with a particular orbital energy.—a spatial distribution of electrons in a molecule that is associated with a particular orbital energy. As the name suggests, molecular orbitals are not localized on a single atom but extend over the entire molecule. Consequently, the molecular orbital approach, called molecular orbital theory is a delocalized approach to bonding. Although the molecular orbital theory is computationally demanding, the principles on which it is based are similar to those we used to determine electron configurations for atoms. -

Atomic Structures of the M Olecular Components in DNA and RNA Based on Bond Lengths As Sums of Atomic Radii
1 Atomic St ructures of the M olecular Components in DNA and RNA based on Bond L engths as Sums of Atomic Radii Raji Heyrovská (Institute of Biophysics, Academy of Sciences of the Czech Republic) E-mail: [email protected] Abstract The author’s interpretation in recent years of bond lengths as sums of the relevant atomic/ionic radii has been extended here to the bonds in the skeletal structures of adenine, guanine, thymine, cytosine, uracil, ribose, deoxyribose and phosphoric acid. On examining the bond length data in the literature, it has been found that the averages of the bond lengths are close to the sums of the corresponding atomic covalent radii of carbon (tetravalent), nitrogen (trivalent), oxygen (divalent), hydrogen (monovalent) and phosphorus (pentavalent). Thus, the conventional molecular structures have been resolved here (for the first time) into probable atomic structures. 1. Introduction The interpretation of the bond lengths in the molecules constituting DNA and RNA have been of interest ever since the discovery [1] of the molecular structure of nucleic acids. This work was motivated by the earlier findings [2a,b] that the inter-atomic distance between any two atoms or ions is, in general, the sum of the radii of the atoms (and or ions) constituting the chemical bond, whether the bond is partially or fully ionic or covalent. It 2 was shown in [3] that the lengths of the hydrogen bonds in the Watson- Crick [1] base pairs (A, T and C, G) of nucleic acids as well as in many other inorganic and biochemical compounds are sums of the ionic or covalent radii of the hydrogen donor and acceptor atoms or ions and of the transient proton involved in the hydrogen bonding. -

Bond Distances and Bond Orders in Binuclear Metal Complexes of the First Row Transition Metals Titanium Through Zinc
Metal-Metal (MM) Bond Distances and Bond Orders in Binuclear Metal Complexes of the First Row Transition Metals Titanium Through Zinc Richard H. Duncan Lyngdoh*,a, Henry F. Schaefer III*,b and R. Bruce King*,b a Department of Chemistry, North-Eastern Hill University, Shillong 793022, India B Centre for Computational Quantum Chemistry, University of Georgia, Athens GA 30602 ABSTRACT: This survey of metal-metal (MM) bond distances in binuclear complexes of the first row 3d-block elements reviews experimental and computational research on a wide range of such systems. The metals surveyed are titanium, vanadium, chromium, manganese, iron, cobalt, nickel, copper, and zinc, representing the only comprehensive presentation of such results to date. Factors impacting MM bond lengths that are discussed here include (a) n+ the formal MM bond order, (b) size of the metal ion present in the bimetallic core (M2) , (c) the metal oxidation state, (d) effects of ligand basicity, coordination mode and number, and (e) steric effects of bulky ligands. Correlations between experimental and computational findings are examined wherever possible, often yielding good agreement for MM bond lengths. The formal bond order provides a key basis for assessing experimental and computationally derived MM bond lengths. The effects of change in the metal upon MM bond length ranges in binuclear complexes suggest trends for single, double, triple, and quadruple MM bonds which are related to the available information on metal atomic radii. It emerges that while specific factors for a limited range of complexes are found to have their expected impact in many cases, the assessment of the net effect of these factors is challenging. -

Calculating Lattice Energies Using the Born-Haber Cycle an Extension Activity for AP Chemistry Students
Calculating Lattice Energies Using the Born-Haber Cycle An Extension Activity for AP Chemistry Students A particular set of equations known as a Born-Haber cycle demonstrates how chemists are able to use the first law of thermodynamics—that the energy of the universe is conserved in any chemical or physical change—to find an unknown energy value that is difficult or impossible to measure experimentally. Some energy quantities, such as the lattice energy of a mineral or the electron affinity of an atom, can be difficult to measure in the lab. Examining a Born-Haber cycle we see that there is more than one path to the formation of a substance in a particular state, and that if we use consistent definitions, an energy value that we seek can be calculated from energy values that we already know. The following exercise will help us see the way these energy values relate to one another, give us practice with their definitions and symbols, and deepen our insight to their meaning when we see them in other types of problems. Each physical or chemical change represented has: • an equation that represents a clearly defined physical or chemical change; • a definition of the particular type of energy change; • a symbol or abbreviation for the energy change equal to a value for the change in enthalpy (ΔH), the energy that is released or absorbed during the change, expressed in kJ/mol; and • a name by which that change in enthalpy is commonly known. To set up the equations in a Born-Haber cycle, cut out the cards for names, equations, defini- tions, and symbols with energy values. -

The Theoretical Study on Interaction of Hydrogen with Single-Walled Boron Nitride Nanotubes
THE JOURNAL OF CHEMICAL PHYSICS 123, 114703 ͑2005͒ The theoretical study on interaction of hydrogen with single-walled boron nitride nanotubes. I. The reactive force field ReaxFFHBN development ͒ Sang Soo Han, Jeung Ku Kang, and Hyuck Mo Leea Department of Materials Science and Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon 305-701, Republic of Korea Adri C. T. van Duin and William A. Goddard III Materials and Process Simulation Center, California Institute of Technology, California 91125 ͑Received 2 November 2004; accepted 21 June 2005; published online 19 September 2005͒ We present a new reactive force field ReaxFFHBN derived to accurately model large molecular and condensed phase systems of H, B, and N atoms. ReaxFFHBN has been tested against quantum calculation data for B–H, B–B, and B–N bond dissociations and for H–B–H, B–N–B, and N–B–N bond angle strain energies of various molecular clusters. The accuracy of the developed ReaxFFHBN for B–N–H systems is also tested for ͑i͒ H–B and H–B bond energies as a function of out of plane ͑ ͒ ͑ ͒ ͑ ͒ in H–B NH2 3 and H–N BH2 3, respectively, ii the reaction energy for the B3N3H6 +H2 → ͑ ͒ B3N3H8, and iii crystal properties such as lattice parameters and equations of states for the hexagonal type ͑h-BN͒ with a graphite structure and for the cubic type ͑c-BN͒ with a zinc-blende structure. For all these systems, ReaxFFHBN gives reliable results consistent with those from quantum calculations as it describes well bond breaking and formation in chemical processes and physical properties. -

Session #9: Homework Solutions
Session #9: Homework Solutions Problem #1 Estimate the ionic radius of Cs+. The lattice energy of CsCl is 633 kJ/mol. For CsCl the Madelung constant, M, is 1.763, and the Born exponent, n, is 10.7. The ionic radius of Cl– is known to be 1.81 Å. Solution Mq q ⎛⎞1 E1 = 1 2 −= and r r+r lattice ⎜⎟ o Cs+ Cl− 4rπεoo⎝⎠ n Solve first for ro MqqN ⎛⎞1 1.763(1.6 x 10-19 ) 2 6.02 x 1023 ⎛ 1 ⎞ r1 = 12 Aν −− = 1 o ⎜⎟ -12 5 ⎜ ⎟ 4Eπεolattice⎝⎠ n 4π 8.85 x 10 6.33x10 ⎝10.7⎠ = 3.50 x 10-10 m = 3.50 Å = r+ r Cs+- Cl ∴ r = 3.50-1.81 = 1.69 Å Cs+ Problem #2 (a) CFCs have been implicated in ozone depletion. Show that when Freon 12 (CCl2F2) is exposed to ultraviolet radiation, the compound decomposes to produce chlorine. (b) Draw the Lewis structure of Freon 12 and indicate the polarities of each bond within this compound. (c) Determine the percent ionic character of the C--Cl and C--F bonds. DATA: Average Bond Energies (kJ/mol) Single Bonds Multiple Bonds H-H 435 C = C 610 F-F 155 C ≡ C 836 Cl-Cl 242 C-C 347 Solution (a) Chlorine will be liberated if the C-Cl bond breaks, therefore, compute its strength and show that ultraviolet photons have enough energy to break the bond. 2 ECCl−−− =+ E CCx9 E ClCl 6.3(χχC− Cl) = 347 x 242 + 96.3(2.55 - 3.16)2 = 326 x 103/6.02 x 1023 = 5.41 x 10-19 J/bond photon will break ∴this bond if Eph > Ebond ⇒ critical λ is hc 6.6 x 10-34 x 3 x 108 λ = = = 3.66 x 10-7 m -19 Ebond 5.41 x 10 which lies in the u.v. -

Real Space in Situ Bond Energies: Toward a Consistent
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Repositorio Institucional de la Universidad de Oviedo Real space in situ bond energies: toward a consistent energetic definition of bond strength D. Menéndez-Crespo, A. Costales, E. Francisco, and A. Martín Pendása∗ June 20, 2018 Abstract A rigorous definition of intrinsic bond strength based on the partitioning of a molecule into real space fragments is presented. Using the domains provided by the quantum theory of atoms in molecules (QTAIM) together with the interacting quantum atoms (IQA) energetic decomposition, we show how an in situ bond strength, matching all the requirements of an intrinsic bond energy, can be defined between each pair of fragments. Total atomization or fragmentation energies are shown to be equal to the sum of these in situ bond energies (ISBEs) if the energies of the fragments are measured with respect to their in-the-molecule state. These energies usually lie above the ground state of the isolated fragments by quantities identified with the standard frag- ment relaxation or deformation energies, which are also provided by the protocol. Deformation energies bridge dissociation energies with ISBEs, and can be dissected using well-known tools of real space theories of chemical bonding. Similarly, ISBEs can be partitioned into ionic and covalent contributions, and this feature adds to the chemical appeal of the procedure. All the energetic quantities examined are observable and amenable, in principle, to experimental deter- mination. Several systems, exemplifying the role of each energetic term herein presented are used to show the power of the approach. -

Bond-Length Distributions for Ions Bonded to Oxygen: Results for the Lanthanides and Actinides and Discussion of the F-Block Contraction
To appear in Acta Crystallographica Section B Bond-length distributions for ions bonded to oxygen: Results for the lanthanides and actinides and discussion of the f-block contraction Olivier Charles Gagnéa* aGeological Sciences, University of Manitoba, 125 Dysart Road, Winnipeg, Manitoba, R3T 2N2, Canada Correspondence email: [email protected] Synopsis Bond-length distributions have been examined for eighty-four configurations of the lanthanide ions and twenty-two configurations of the actinide ions bonded to oxygen. The lanthanide contraction for the trivalent lanthanide ions bonded to O2- is shown to vary as a function of coordination number and to diminish in scale with increasing coordination number. Abstract Bond-length distributions have been examined for eighty-four configurations of the lanthanide ions and twenty-two configurations of the actinide ions bonded to oxygen, for 1317 coordination polyhedra and 10,700 bond distances for the lanthanide ions, and 671 coordination polyhedra and 4754 bond distances for the actinide ions. A linear correlation between mean bond-length and coordination number is observed for the trivalent lanthanides ions bonded to O2-. The lanthanide contraction for the trivalent lanthanide ions bonded to O2- is shown to vary as a function of coordination number, and to diminish in scale with an increasing coordination number. The decrease in mean bond- length from La3+ to Lu3+ is 0.25 Å for CN 6 (9.8%), 0.22 Å for CN 7 (8.7%), 0.21 Å for CN 8 (8.0%), 0.21 Å for CN 9 (8.2%) and 0.18 Å for CN 10 (6.9%). The crystal chemistry of Np5+ and Np6+ is shown to be very similar to that of U6+ when bonded to O2-, but differs for Np7+. -

Chapter 5 the Covalent Bond 1
Chapter 5 The Covalent Bond 1. What two opposing forces dictate the bond length? (Why do bonds form and what keeps the bonds from getting any shorter?) The bond length is the distance at which the repulsion of the two nuclei equals the attraction of the valence electrons on one atom and the nucleus of the other. 3. List the following bonds in order of increasing bond length: H-Cl, H-Br, H-O. H-O < H-Cl < H-Br. The order of increasing atom size. 5. Use electronegativities to describe the nature (purely covalent, mostly covalent, polar covalent, or ionic) of the following bonds. a) P-Cl Δχ = 3.0 – 2.1 = 0.9 polar covalent b) K-O Δχ = 3.5 – 0.8 = 2.7 ionic c) N-H Δχ = 3.0 – 2.1 = 0.9 polar covalent d) Tl-Br Δχ = 2.8 – 1.7 = 1.1 polar covalent 7. Name the following compounds: a) S2Cl2 disulfur dichloride b) CCl4 carbon tetrachloride c) PCl5 phosphorus pentachloride d) HCl hydrogen chloride 9. Write formulas for each of the following compounds: a) dinitrogen tetroxide N2O4 b) nitrogen monoxide NO c) dinitrogen pentoxide N2O5 11. Consider the U-V, W-X, and Y-Z bonds. The valence orbital diagrams for the orbitals involved in the bonds are shown in the margin. Use an arrow to show the direction of the bond dipole in the polar bonds or indicate “not polar” for the nonpolar bond(s). Rank the bonds in order of increasing polarity. The bond dipole points toward the more electronegative atom in the bond, which is the atom with the lower energy orbital.