Lawrence Berkeley National Laboratory Recent Work
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Vertex Coloring
Lecture 1 Vertex Coloring 1.1 The Problem Nowadays multi-core computers get more and more processors, and the question is how to handle all this parallelism well. So, here's a basic problem: Consider a doubly linked list that is shared by many processors. It supports insertions and deletions, and there are simple operations like summing up the size of the entries that should be done very fast. We decide to organize the data structure as an array of dynamic length, where each array index may or may not hold an entry. Each entry consists of the array indices of the next entry and the previous entry in the list, some basic information about the entry (e.g. its size), and a pointer to the lion's share of the data, which can be anywhere in the memory. Memory structure Logical structure 1 2 3 4 5 6 7 8 9 1 2 3 4 5 6 7 8 9 next previous size data Figure 1.1: Linked listed after initialization. Blue links are forward pointers, red links backward pointers (these are omitted from now on). We now can quickly determine the total size by reading the array in one go from memory, which is quite fast. However, how can we do insertions and deletions fast? These are local operations affecting only one list entry and the pointers of its \neighbors," i.e., the previous and next list element. We want to be able to do many such operations concurrently, by different processors, while maintaining the link structure! Being careless and letting each processor act independently invites desaster, see Figure 1.3. -

Easy PRAM-Based High-Performance Parallel Programming with ICE∗
Easy PRAM-based high-performance parallel programming with ICE∗ Fady Ghanim1, Uzi Vishkin1,2, and Rajeev Barua1 1Electrical and Computer Engineering Department 2University of Maryland Institute for Advance Computer Studies University of Maryland - College Park MD, 20742, USA ffghanim,barua,[email protected] Abstract Parallel machines have become more widely used. Unfortunately parallel programming technologies have advanced at a much slower pace except for regular programs. For irregular programs, this advancement is inhibited by high synchronization costs, non-loop parallelism, non-array data structures, recursively expressed parallelism and parallelism that is too fine-grained to be exploitable. We present ICE, a new parallel programming language that is easy-to-program, since: (i) ICE is a synchronous, lock-step language; (ii) for a PRAM algorithm its ICE program amounts to directly transcribing it; and (iii) the PRAM algorithmic theory offers unique wealth of parallel algorithms and techniques. We propose ICE to be a part of an ecosystem consisting of the XMT architecture, the PRAM algorithmic model, and ICE itself, that together deliver on the twin goal of easy programming and efficient parallelization of irregular programs. The XMT architecture, developed at UMD, can exploit fine-grained parallelism in irregular programs. We built the ICE compiler which translates the ICE language into the multithreaded XMTC language; the significance of this is that multi-threading is a feature shared by practically all current scalable parallel programming languages. Our main result is perhaps surprising: The run-time was comparable to XMTC with a 0.48% average gain for ICE across all benchmarks. Also, as an indication of ease of programming, we observed a reduction in code size in 7 out of 11 benchmarks vs. -

Instructor's Manual
Instructor’s Manual Vol. 2: Presentation Material CCC Mesh/Torus Butterfly !*#? Sea Sick Hypercube Pyramid Behrooz Parhami This instructor’s manual is for Introduction to Parallel Processing: Algorithms and Architectures, by Behrooz Parhami, Plenum Series in Computer Science (ISBN 0-306-45970-1, QA76.58.P3798) 1999 Plenum Press, New York (http://www.plenum.com) All rights reserved for the author. No part of this instructor’s manual may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, microfilming, recording, or otherwise, without written permission. Contact the author at: ECE Dept., Univ. of California, Santa Barbara, CA 93106-9560, USA ([email protected]) Introduction to Parallel Processing: Algorithms and Architectures Instructor’s Manual, Vol. 2 (4/00), Page iv Preface to the Instructor’s Manual This instructor’s manual consists of two volumes. Volume 1 presents solutions to selected problems and includes additional problems (many with solutions) that did not make the cut for inclusion in the text Introduction to Parallel Processing: Algorithms and Architectures (Plenum Press, 1999) or that were designed after the book went to print. It also contains corrections and additions to the text, as well as other teaching aids. The spring 2000 edition of Volume 1 consists of the following parts (the next edition is planned for spring 2001): Vol. 1: Problem Solutions Part I Selected Solutions and Additional Problems Part II Question Bank, Assignments, and Projects Part III Additions, Corrections, and Other Updates Part IV Sample Course Outline, Calendar, and Forms Volume 2 contains enlarged versions of the figures and tables in the text, in a format suitable for use as transparency masters. -
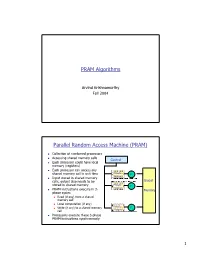
PRAM Algorithms Parallel Random Access Machine
PRAM Algorithms Arvind Krishnamurthy Fall 2004 Parallel Random Access Machine (PRAM) n Collection of numbered processors n Accessing shared memory cells Control n Each processor could have local memory (registers) n Each processor can access any shared memory cell in unit time Private P Memory 1 n Input stored in shared memory cells, output also needs to be Global stored in shared memory Private P Memory 2 n PRAM instructions execute in 3- Memory phase cycles n Read (if any) from a shared memory cell n Local computation (if any) Private n Write (if any) to a shared memory P Memory p cell n Processors execute these 3-phase PRAM instructions synchronously 1 Shared Memory Access Conflicts n Different variations: n Exclusive Read Exclusive Write (EREW) PRAM: no two processors are allowed to read or write the same shared memory cell simultaneously n Concurrent Read Exclusive Write (CREW): simultaneous read allowed, but only one processor can write n Concurrent Read Concurrent Write (CRCW) n Concurrent writes: n Priority CRCW: processors assigned fixed distinct priorities, highest priority wins n Arbitrary CRCW: one randomly chosen write wins n Common CRCW: all processors are allowed to complete write if and only if all the values to be written are equal A Basic PRAM Algorithm n Let there be “n” processors and “2n” inputs n PRAM model: EREW n Construct a tournament where values are compared P0 Processor k is active in step j v if (k % 2j) == 0 At each step: P0 P4 Compare two inputs, Take max of inputs, P0 P2 P4 P6 Write result into shared memory -

CSE 4351/5351 Notes 9: PRAM and Other Theoretical Model S
CSE 4351/5351 Notes 9: PRAM and Other Theoretical Model s Shared Memory Model Traditional Sequential Algorithm Model RAM (Random Access Machine) Uniform access time to memory Arithmetic operations performed in O(1) time (simplification, really O(lg n)) Generalization of RAM to Parallel Computation PRAM (Parallel Random Access Machine) SHARED MEMORY P0 P1 P2 P3 . Pn FAMILY of models - different concurrency assumptions imply DRASTIC differences in hardware/software support or may be impossible to physically realize. MAJOR THEORETICAL ISSUE: What is the penalty for modifying an algorithm in a flexible model to satisfy a restrictive model? MAJOR PRACTICAL ISSUES: Adequacy for mapping to popular topologies. Developing topologies/algorithms to support models with minimum penalty. Other models: Valiant's BSP (bulk-synchronous parallel), CACM 33 (8), August 1990 "Large" supersteps Bulk communication/routing between supersteps Syncronization to determine superstep completion Karp's LogP, ACM Symp. on Parallel Algorithms and Architectures, 1993 Processors can be: Operational: Computing, receiving, submitting a message Stalling: Delay in message being accepted by communication medium Parameters: L: Maximum time for message to arrive o: Overhead for preparing each message g: Time units between consecutive communication handling steps P: Maximum processor computation time PRAM algorithms are usually described in a SIMD fashion 2 Highly synchronized by constructs in programming notation Instructions are modified based on the processor id Algorithms are simplified by letting processors do useless work Lends to data parallel implementation - PROGRAMMER MUST REDUCE SYNCHRONIZATION OVERHEAD Example PRAM Models EREW (Exclusive Read, Exclusive Write) - Processors must access different memory cells. Most restrictive. CREW (Concurrent Read, Exclusive Write) - Either multiple reads or a single writer may access a cell ERCW (Exclusive Read, Concurrent Write) - Not popular CRCW (Concurrent Read, Concurrent Write) - Most flexible. -

Towards a Spatial Model Checker on GPU⋆
Towards a spatial model checker on GPU? Laura Bussi1, Vincenzo Ciancia2, and Fabio Gadducci1 1 Dipartimento di Informatica, Universit`adi Pisa 2 Istituto di Scienza e Tecnologie dell'Informazione, CNR Abstract. The tool VoxLogicA merges the state-of-the-art library of computational imaging algorithms ITK with the combination of declara- tive specification and optimised execution provided by spatial logic model checking. The analysis of an existing benchmark for segmentation of brain tumours via a simple logical specification reached very high accu- racy. We introduce a new, GPU-based version of VoxLogicA and present preliminary results on its implementation, scalability, and applications. Keywords: Spatial logics · Model Checking · GPU computation 1 Introduction and background Spatial and Spatio-temporal model checking have gained an increasing interest in recent years in various application domains, including collective adaptive [12, 11] and networked systems [5], runtime monitoring [17, 15, 4], modelling of cyber- physical systems [20] and medical imaging [13, 3]. Introduced in [7], VoxLogicA (Voxel-based Logical Analyser)3 caters for a declarative approach to (medical) image segmentation, supported by spatial model checking. A spatial logic is defined, tailored to high-level imaging features, such as regions, contact, texture, proximity, distance. Spatial operators are mostly derived from the Spatial Logic of Closure Spaces (SLCS, see Figure 1). Models of the spatial logic are (pixels of) images, with atomic propositions given by imaging features (e.g. colour, intensity), and spatial structure obtained via adjacency of pixels. SLCS features a modal operator near, denoting adjacency of pixels, and a reachability operator ρ φ1[φ2], holding at pixel x whenever there is a path from x to a pixel y satisfying φ1, with all intermediate points, except the extremes, satisfying φ2. -

2009-04-21 AMSC Cloud Assembler
Towards a de novo short read assembler for large genomes using cloud computing Michael Schatz April 21, 2009 AMSC664 Advanced Scientific Computing Outline 1.! Genome assembly by analogy 2.! DNA sequencing and assembly 3.! MapReduce for genome assembly 4.! Research plan and related work Shredded Book Reconstruction •! Dickens accidently shreds the original A Tale of Two Cities –! Text printed on 5 long spools It was theIt was best the of besttimes, of times, it was it wasthe worstthe worst of of times, times, it it was was the the ageage of of wisdom, wisdom, it itwas was the agethe ofage foolishness, of foolishness, … … It was theIt was best the bestof times, of times, it was it was the the worst of times, it was the theage ageof wisdom, of wisdom, it was it thewas age the of foolishness,age of foolishness, … It was theIt was best the bestof times, of times, it was it wasthe the worst worst of times,of times, it it was the age of wisdom, it wasit was the the age age of offoolishness, foolishness, … … It was It thewas best the ofbest times, of times, it wasit was the the worst worst of times,of times, itit waswas thethe ageage ofof wisdom,wisdom, it wasit was the the age age of foolishness,of foolishness, … … It wasIt thewas best the bestof times, of times, it wasit was the the worst worst of of times, it was the age of ofwisdom, wisdom, it wasit was the the age ofage foolishness, of foolishness, … … •! How can he reconstruct the text? –! 5 copies x 138, 656 words / 5 words per fragment = 138k fragments –! The short fragments from every -

Implementing a Tool for Designing Portable Parallel Programs
Copyright Warning & Restrictions The copyright law of the United States (Title 17, United States Code) governs the making of photocopies or other reproductions of copyrighted material. Under certain conditions specified in the law, libraries and archives are authorized to furnish a photocopy or other reproduction. One of these specified conditions is that the photocopy or reproduction is not to be “used for any purpose other than private study, scholarship, or research.” If a, user makes a request for, or later uses, a photocopy or reproduction for purposes in excess of “fair use” that user may be liable for copyright infringement, This institution reserves the right to refuse to accept a copying order if, in its judgment, fulfillment of the order would involve violation of copyright law. Please Note: The author retains the copyright while the New Jersey Institute of Technology reserves the right to distribute this thesis or dissertation Printing note: If you do not wish to print this page, then select “Pages from: first page # to: last page #” on the print dialog screen The Van Houten library has removed some of the personal information and all signatures from the approval page and biographical sketches of theses and dissertations in order to protect the identity of NJIT graduates and faculty. ABSTRACT Implementing A Tool For Designing Portable Parallel Programs by Geetha Chitti The Implementation aspects of a novel parallel pro- gramming model called Cluster-M is presented in this the- sis. This model provides an environment for efficiently de- signing highly parallel portable software. The two main components of this model are Cluster-M Specifications and Cluster-M Representations. -

Models for Parallel Computation in Multi-Core, Heterogeneous, and Ultra Wide-Word Architectures
Models for Parallel Computation in Multi-Core, Heterogeneous, and Ultra Wide-Word Architectures by Alejandro Salinger A thesis presented to the University of Waterloo in fulfillment of the thesis requirement for the degree of Doctor of Philosophy in Computer Science Waterloo, Ontario, Canada, 2013 c Alejandro Salinger 2013 I hereby declare that I am the sole author of this thesis. This is a true copy of the thesis, including any required final revisions, as accepted by my examiners. I understand that my thesis may be made electronically available to the public. iii Abstract Multi-core processors have become the dominant processor architecture with 2, 4, and 8 cores on a chip being widely available and an increasing number of cores predicted for the future. In ad- dition, the decreasing costs and increasing programmability of Graphic Processing Units (GPUs) have made these an accessible source of parallel processing power in general purpose computing. Among the many research challenges that this scenario has raised are the fundamental problems related to theoretical modeling of computation in these architectures. In this thesis we study several aspects of computation in modern parallel architectures, from modeling of computation in multi-cores and heterogeneous platforms, to multi-core cache management strategies, through the proposal of an architecture that exploits bit-parallelism on thousands of bits. Observing that in practice multi-cores have a small number of cores, we propose a model for low-degree parallelism for these architectures. We argue that assuming a small number of processors (logarithmic in a problem's input size) simplifies the design of parallel algorithms. -

PRAM Algorithms Parallel Random Access Machine
Parallel Random Access Machine (PRAM) n Collection of numbered processors n Accessing shared memory cells Control n Each processor could have local memory (registers) PRAM Algorithms n Each processor can access any shared memory cell in unit time Private P Memory 1 n Input stored in shared memory cells, output also needs to be Global stored in shared memory Private P Memory 2 Arvind Krishnamurthy n PRAM instructions execute in 3- Memory phase cycles Fall 2004 n Read (if any) from a shared memory cell n Local computation (if any) Private n Write (if any) to a shared memory P Memory p cell n Processors execute these 3-phase PRAM instructions synchronously Shared Memory Access Conflicts A Basic PRAM Algorithm n Different variations: n Let there be “n” processors and “2n” inputs n Exclusive Read Exclusive Write (EREW) PRAM: no two processors n PRAM model: EREW are allowed to read or write the same shared memory cell simultaneously n Construct a tournament where values are compared n Concurrent Read Exclusive Write (CREW): simultaneous read allowed, but only one processor can write P0 Processor k is active in step j v if (k % 2j) == 0 n Concurrent Read Concurrent Write (CRCW) At each step: n Concurrent writes: P0 P4 Compare two inputs, n Priority CRCW: processors assigned fixed distinct priorities, highest Take max of inputs, priority wins P0 P2 P4 P6 Write result into shared memory n Arbitrary CRCW: one randomly chosen write wins P0 P1 P2 P3 P4 P5 P6 P7 Details: n Common CRCW: all processors are allowed to complete write if and only if all -

Introduction to Parallel Processing : Algorithms and Architectures
Introduction to Parallel Processing Algorithms and Architectures PLENUM SERIES IN COMPUTER SCIENCE Series Editor: Rami G. Melhem University of Pittsburgh Pittsburgh, Pennsylvania FUNDAMENTALS OF X PROGRAMMING Graphical User Interfaces and Beyond Theo Pavlidis INTRODUCTION TO PARALLEL PROCESSING Algorithms and Architectures Behrooz Parhami Introduction to Parallel Processing Algorithms and Architectures Behrooz Parhami University of California at Santa Barbara Santa Barbara, California KLUWER ACADEMIC PUBLISHERS NEW YORK, BOSTON , DORDRECHT, LONDON , MOSCOW eBook ISBN 0-306-46964-2 Print ISBN 0-306-45970-1 ©2002 Kluwer Academic Publishers New York, Boston, Dordrecht, London, Moscow All rights reserved No part of this eBook may be reproduced or transmitted in any form or by any means, electronic, mechanical, recording, or otherwise, without written consent from the Publisher Created in the United States of America Visit Kluwer Online at: http://www.kluweronline.com and Kluwer's eBookstore at: http://www.ebooks.kluweronline.com To the four parallel joys in my life, for their love and support. This page intentionally left blank. Preface THE CONTEXT OF PARALLEL PROCESSING The field of digital computer architecture has grown explosively in the past two decades. Through a steady stream of experimental research, tool-building efforts, and theoretical studies, the design of an instruction-set architecture, once considered an art, has been transformed into one of the most quantitative branches of computer technology. At the same time, better understanding of various forms of concurrency, from standard pipelining to massive parallelism, and invention of architectural structures to support a reasonably efficient and user-friendly programming model for such systems, has allowed hardware performance to continue its exponential growth. -

Parallel Peak Pruning for Scalable SMP Contour Tree Computation
Lawrence Berkeley National Laboratory Recent Work Title Parallel peak pruning for scalable SMP contour tree computation Permalink https://escholarship.org/uc/item/659949x4 ISBN 9781509056590 Authors Carr, HA Weber, GH Sewell, CM et al. Publication Date 2017-03-08 DOI 10.1109/LDAV.2016.7874312 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Parallel Peak Pruning for Scalable SMP Contour Tree Computation Hamish A. Carr∗ Gunther H. Webery Christopher M. Sewellz James P. Ahrensx University of Leeds Lawrence Berkeley National Laboratory Los Alamos National Laboratory University of California, Davis ABSTRACT output to disk—as does the recognition that one component of As data sets grow to exascale, automated data analysis and visu- the pipeline remains unchanged: the human perceptual system. alisation are increasingly important, to intermediate human under- This need has stimulated research into areas such as computational standing and to reduce demands on disk storage via in situ anal- topology, which constructs models of the mathematical structure ysis. Trends in architecture of high performance computing sys- of the data for the purposes of analysis and visualisation. One of tems necessitate analysis algorithms to make effective use of com- the principal mathematical tools is the contour tree or Reeb graph, binations of massively multicore and distributed systems. One of which summarizes the development of contours in the data set as the principal analytic tools is the contour tree, which analyses rela- the isovalue varies. Since contours are a key element of most visu- tionships between contours to identify features of more than local alisations, the contour tree and the related merge tree are of prime importance.