Accession Number Description Fold Enrichment Uniquepeptidecount
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Kinesin-14 Motor Protein KIFC1 Participates in DNA Synthesis and Chromatin Maintenance Ya-Lan Wei1 and Wan-Xi Yang 1
Wei and Yang Cell Death and Disease (2019) 10:402 https://doi.org/10.1038/s41419-019-1619-9 Cell Death & Disease ARTICLE Open Access Kinesin-14 motor protein KIFC1 participates in DNA synthesis and chromatin maintenance Ya-Lan Wei1 and Wan-Xi Yang 1 Abstract The nuclear localization signal (NLS) in kinesin-14 KIFC1 is associated with nuclear importins and Ran gradient, but detailed mechanism remains unknown. In this study, we found that KIFC1 proteins have specific transport characteristics during cell cycle. In the absence of KIFC1, cell cycle kinetics decrease significantly with a prolonged S phase. After KIFC1 overexpression, the duration of S phase becomes shorten. KIFC1 may transport the recombinant/ replicate-related proteins into the nucleus, meanwhile avoiding excessive KIFC1 in the cytoplasm, which results in aberrant microtubule bundling. Interestingly, the deletion of kifc1 in human cells results in a higher ratio of aberrant nuclear membrane, and the degradation of lamin B and lamin A/C. We also found that kifc1 deletion leads to defects in metaphase mitotic spindle assembly, and then results in chromosome structural abnormality. The kifc1-/- cells finally form micronuclei in daughter cells, and results in aneuploidy and chromosome loss in cell cycle. In this study, we demonstrate that kinesin-14 KIFC1 proteins involve in regulating DNA synthesis in S phase, and chromatin maintenance in mitosis, and maintain cell growth in a nuclear transport-independent way. 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; Introduction KIFC1 mainly cluster the spindles involving in chromo- Kinesin-14 KIFC1 transports various cargos along the some alignment and segregation. While chromokinesins microtubule to the minus ends1. -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

Plasma Membrane Ca2+–Atpase in Rat and Human Odontoblasts Mediates Dentin Mineralization
biomolecules Article Plasma Membrane Ca2+–ATPase in Rat and Human Odontoblasts Mediates Dentin Mineralization Maki Kimura 1,†, Hiroyuki Mochizuki 1,†, Ryouichi Satou 2, Miyu Iwasaki 2, Eitoyo Kokubu 3, Kyosuke Kono 1, Sachie Nomura 1, Takeshi Sakurai 1, Hidetaka Kuroda 1,4,† and Yoshiyuki Shibukawa 1,*,† 1 Department of Physiology, Tokyo Dental College, 2-9-18, Kanda-Misaki-cho, Chiyoda-ku, Tokyo 101-0061, Japan; [email protected] (M.K.); [email protected] (H.M.); [email protected] (K.K.); [email protected] (S.N.); [email protected] (T.S.); [email protected] (H.K.) 2 Department of Epidemiology and Public Health, Tokyo Dental College, Chiyodaku, Tokyo 101-0061, Japan; [email protected] (R.S.); [email protected] (M.I.) 3 Department of Microbiology, Tokyo Dental College, Chiyodaku, Tokyo 101-0061, Japan; [email protected] 4 Department of Dental Anesthesiology, Kanagawa Dental University, 1-23, Ogawacho, Kanagawa, Yokosuka-shi 238-8570, Japan * Correspondence: [email protected] † These authors contributed equally to this study. Abstract: Intracellular Ca2+ signaling engendered by Ca2+ influx and mobilization in odontoblasts is critical for dentinogenesis induced by multiple stimuli at the dentin surface. Increased Ca2+ is exported by the Na+–Ca2+ exchanger (NCX) and plasma membrane Ca2+–ATPase (PMCA) to Citation: Kimura, M.; Mochizuki, H.; maintain Ca2+ homeostasis. We previously demonstrated a functional coupling between Ca2+ Satou, R.; Iwasaki, M.; Kokubu, E.; extrusion by NCX and its influx through transient receptor potential channels in odontoblasts. Kono, K.; Nomura, S.; Sakurai, T.; Although the presence of PMCA in odontoblasts has been previously described, steady-state levels of Kuroda, H.; Shibukawa, Y. -

Kinesin Family Member 18B Regulates the Proliferation and Invasion Of
Wu et al. Cell Death and Disease (2021) 12:302 https://doi.org/10.1038/s41419-021-03582-2 Cell Death & Disease ARTICLE Open Access Kinesin family member 18B regulates the proliferation and invasion of human prostate cancer cells Yu-Peng Wu 1,Zhi-BinKe 1, Wen-Cai Zheng 1, Ye-Hui Chen 1,Jun-MingZhu 1,FeiLin 1,Xiao-DongLi 1, Shao-Hao Chen 1,HaiCai 1, Qing-Shui Zheng 1, Yong Wei 1, Xue-Yi Xue 1 and Ning Xu 1 Abstract Expression of kinesin family member 18B (KIF18B), an ATPase with key roles in cell division, is deregulated in many cancers, but its involvement in prostate cancer (PCa) is unclear. Here, we investigated the expression and function of KIF18B in human PCa specimens and cell lines using bioinformatics analyses, immunohistochemical and immunofluorescence microscopy, and RT-qPCR and western blot analyses. KIF18B was overexpressed in PCa specimens compared with paracancerous tissues and was associated with poorer disease-free survival. In vitro, KIF18B knockdown in PCa cell lines promoted cell proliferation, migration, and invasion, and inhibited cell apoptosis, while KIF18B overexpression had the opposite effects. In a mouse xenograft model, KIF18B overexpression accelerated and promoted the growth of PCa tumors. Bioinformatics analysis of control and KIF18B-overexpressing PCa cells showed that genes involved in the PI3K–AKT–mTOR signaling pathway were significantly enriched among the differentially expressed genes. Consistent with this observation, we found that KIF18B overexpression activates the PI3K–AKT–mTOR signaling pathway in PCa cells both in vitro and in vivo. Collectively, our results suggest that KIF18B plays a crucial role – – 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; in PCa via activation of the PI3K AKT mTOR signaling pathway, and raise the possibility that KIF18B could have utility as a novel biomarker for PCa. -

Electronic Supplementary Material (ESI) for Molecular Biosystems
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is © The Royal Society of Chemistry 2015 Table S5 Mass data of proteins identified with label free shotgun proteomics a #Alt. Proteins; b Scores; c #Peptides; d SC [%]; e RMS90 [ppm]; f Rank; g Median(Controls:GLP1); h #(Controls:GLP1); i CV [%](Controls:GLP1); l Median(Controls:Palmitate); m #(Controls:Palmitate) ; n CV [%](Controls:Palmitate); o Median(Controls:GLP1 + Palmitate); p #(Controls:GLP1 + Palmitate); q CV [%](Controls:GLP1 + Palmitate) MW OK Accession Protein pI a b c d e f g h i l m n o p q [kDa] 78 kDa glucose-regulated protein OS=Rattus 1672.6 true GRP78_RAT 72.3 4.9 1 22 39 1.48 1 1.16 6 20.53 1.18 9 9.56 1.24 10 13.52 norvegicus GN=Hspa5 PE=1 SV=1 (M:1672.6) Endoplasmin OS=Rattus norvegicus 1253.0 true ENPL_RAT 92.7 4.6 1 23 29 3.34 2 0.85 8 33.34 1.19 6 39.58 1.18 10 28.11 GN=Hsp90b1 PE=1 SV=2 (M:1253.0) Stress-70 protein, mitochondrial OS=Rattus 1138.8 true GRP75_RAT 73.8 5.9 1 17 28.7 1.18 3 1.21 5 7.62 0.78 1 1.1 8 8.04 norvegicus GN=Hspa9 PE=1 SV=3 (M:1138.8) Protein disulfide-isomerase A3 OS=Rattus 1035.4 true PDIA3_RAT 56.6 5.8 1 16 35.2 2.84 4 0.94 3 17.22 1.15 4 26.62 1.25 4 7.86 norvegicus GN=Pdia3 PE=1 SV=2 (M:1035.4) Aconitate hydratase, mitochondrial 983.8 true ACON_RAT OS=Rattus norvegicus GN=Aco2 PE=1 85.4 8.7 1 19 31.4 2.72 5 0.89 2 23.6 1.21 2 9.25 0.89 3 32.87 (M:983.8) SV=2 60 kDa heat shock protein, mitochondrial 906.9 true CH60_RAT OS=Rattus norvegicus GN=Hspd1 PE=1 60.9 5.8 1 13 32.5 3.26 6 1.05 5 13.15 1.01 2 32.54 1.01 -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

Molecular Genetics of Microcephaly Primary Hereditary: an Overview
brain sciences Review Molecular Genetics of Microcephaly Primary Hereditary: An Overview Nikistratos Siskos † , Electra Stylianopoulou †, Georgios Skavdis and Maria E. Grigoriou * Department of Molecular Biology & Genetics, Democritus University of Thrace, 68100 Alexandroupolis, Greece; [email protected] (N.S.); [email protected] (E.S.); [email protected] (G.S.) * Correspondence: [email protected] † Equal contribution. Abstract: MicroCephaly Primary Hereditary (MCPH) is a rare congenital neurodevelopmental disorder characterized by a significant reduction of the occipitofrontal head circumference and mild to moderate mental disability. Patients have small brains, though with overall normal architecture; therefore, studying MCPH can reveal not only the pathological mechanisms leading to this condition, but also the mechanisms operating during normal development. MCPH is genetically heterogeneous, with 27 genes listed so far in the Online Mendelian Inheritance in Man (OMIM) database. In this review, we discuss the role of MCPH proteins and delineate the molecular mechanisms and common pathways in which they participate. Keywords: microcephaly; MCPH; MCPH1–MCPH27; molecular genetics; cell cycle 1. Introduction Citation: Siskos, N.; Stylianopoulou, Microcephaly, from the Greek word µικρoκεϕαλi´α (mikrokephalia), meaning small E.; Skavdis, G.; Grigoriou, M.E. head, is a term used to describe a cranium with reduction of the occipitofrontal head circum- Molecular Genetics of Microcephaly ference equal, or more that teo standard deviations -

Murine Neonatal Ketogenesis Preserves Mitochondrial Energetics by Preventing Protein Hyperacetylation
ARTICLES https://doi.org/10.1038/s42255-021-00342-6 Murine neonatal ketogenesis preserves mitochondrial energetics by preventing protein hyperacetylation Yuichiro Arima 1,2,13 ✉ , Yoshiko Nakagawa3,13, Toru Takeo 3,13, Toshifumi Ishida 1, Toshihiro Yamada1, Shinjiro Hino4, Mitsuyoshi Nakao4, Sanshiro Hanada 2, Terumasa Umemoto 2, Toshio Suda2, Tetsushi Sakuma 5, Takashi Yamamoto5, Takehisa Watanabe6, Katsuya Nagaoka6, Yasuhito Tanaka6, Yumiko K. Kawamura7,8, Kazuo Tonami7, Hiroki Kurihara7, Yoshifumi Sato9, Kazuya Yamagata9,10, Taishi Nakamura 1,11, Satoshi Araki1, Eiichiro Yamamoto1, Yasuhiro Izumiya1,12, Kenji Sakamoto1, Koichi Kaikita1, Kenichi Matsushita 1, Koichi Nishiyama2, Naomi Nakagata3 and Kenichi Tsujita1,10 Ketone bodies are generated in the liver and allow for the maintenance of systemic caloric and energy homeostasis during fasting and caloric restriction. It has previously been demonstrated that neonatal ketogenesis is activated independently of starvation. However, the role of ketogenesis during the perinatal period remains unclear. Here, we show that neonatal ketogen- esis plays a protective role in mitochondrial function. We generated a mouse model of insufficient ketogenesis by disrupting the rate-limiting hydroxymethylglutaryl-CoA synthase 2 enzyme gene (Hmgcs2). Hmgcs2 knockout (KO) neonates develop microvesicular steatosis within a few days of birth. Electron microscopic analysis and metabolite profiling indicate a restricted energy production capacity and accumulation of acetyl-CoA in Hmgcs2 KO mice. Furthermore, -

SERCA in Genesis of Arrhythmias: What We Already Know and What Is New?
Review 43 SERCA in genesis of arrhythmias: what we already know and what is new? Nilüfer Erkasap Department of Physiology, Medical Faculty, Eskiflehir Osmangazi University, Eskiflehir, Turkey ABSTRACT This review mainly focuses on the structure, function of the sarco(endo)plasmic reticulum calcium pump (SERCA) and its role in genesis of arrhythmias. SERCA is a membrane protein that belongs to the family of P-type ion translocating ATPases and pumps free cytosolic calcium into intracellular stores. Active transport of Ca2+ is achieved, according to the E1-E2 model, changing of SERCA structure by Ca2+. The affinity of Ca2+ -binding sites varies from high (E1) to low (E2). Three different SERCA genes were identified-SERCA1, SERCA2, and SERCA3. SERCA is mainly represented by the SERCA2a isoform in the heart. In heart muscle, during systole, depolarization triggers the release of Ca2+ from the sarcoplasmic reticulum (SR) and starts contraction. During diastole, muscle relaxation occurs as Ca2+ is again removed from cytosol, predominantly by accumulation into SR via the action of SERCA2a. The main regulator of SERCA2a is phospholamban and another regulator proteolipid of SERCA is sarcolipin. There are a lot of studies on the effect of decreased and/or increased SERCA activity in genesis of arrhythmia. Actually both decrease and increase of SERCA activity in the heart result in some pathological mechanisms such as heart failure and arrhythmia. (Anadolu Kardiyol Derg 2007: 7 Suppl 1; 43-6) Key words: sarco(endo)plasmic reticulum, SERCA, arrhythmia, calcium channels Introduction from cytosol, predominantly by accumulation into sarcoplasmic reticulum via the action of sarco(endo)plasmic reticulum Cardiac physiology is a major area of research in basic and Ca ATPase (SERCA). -

A Common Analgesic Enhances the Anti-Tumour Activity of 5-Aza-2’- Deoxycytidine Through Induction of Oxidative Stress
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.31.017947; this version posted April 1, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A common analgesic enhances the anti-tumour activity of 5-aza-2’- deoxycytidine through induction of oxidative stress Hannah J. Gleneadie1,10, Amy H. Baker1, Nikolaos Batis2, Jennifer Bryant2, Yao Jiang3, Samuel J.H. Clokie4, Hisham Mehanna2, Paloma Garcia5, Deena M.A. Gendoo6, Sally Roberts5, Alfredo A. Molinolo7, J. Silvio Gutkind8, Ben A. Scheven1, Paul R. Cooper1, Farhat L. Khanim9 and Malgorzata Wiench1, 5,*. 1School of Dentistry, Institute of Clinical Studies, College of Medical and Dental Sciences, The University of Birmingham, Birmingham, B5 7EG, UK; 2Institute of Head and Neck Studies and Education (InHANSE), The University of Birmingham, Birmingham, B15 2TT, UK; 3School of Biosciences, The University of Birmingham, Birmingham, B15 2TT, UK; 4West Midlands Regional Genetics Laboratory, Birmingham Women’s and Children’s Hospital, Birmingham, B15 2TG, UK; 5Institute of Cancer and Genomic Sciences, College of Medical and Dental Sciences, The University of Birmingham, Birmingham, B15 2TT, UK; 6Centre for Computational Biology, Institute of Cancer and Genomic Sciences, The University of Birmingham, Birmingham, B15 2TT, UK; 7Moores Cancer Center and Department of Pathology, University of California San Diego, La Jolla, CA 92093, USA; 8Department of Pharmacology and Moores Cancer -
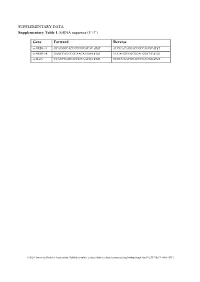
Supplementary Tables and Figures
SUPPLEMENTARY DATA Supplementary Table 1. SiRNA sequence (5’-3’) Gene Forward Reverse si-HRD1-1# GCAUGGCAGUCCUGUACAU dTdT AUGUACAGGACUGCCAUGC dTdT si-HRD1-2# GAGCCAUCCGCAACAUGAA dTdT UUCAUGUUGCGGAUGGCUC dTdT si-MafA CCAUCGAGUACGUCAACGA dTdT UCGUUGACGUACUCGAUGG dTdT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR (5’-3’) Gene Forward Reverse human HRD1 GCTCACGCCTACTACCTCAAA GCCAGACAAGTCTCTGTGACG mouse mafA AAGCGGCGCACGCTCAAGAA GGTCCCGCTCCTTGGCCAGA mouse insulin1 CACTTCCTACCCCTGCTGG ACCACAAAGATGCTGTTTGACA mouse β-actin AGGCCAACCGTGAAAAGATG AGAGCATAGCCCTCGTAGATGG human β-actin CATGTACGTTGCTATCCAGGC CTCCTTAATGTCACGCACGAT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 3. Primer sequences for ChIP (5’-3’) Gene promoter Forward Reverse mouse Insulin1, 2 GGAACTGTGAAACAGTCCAAGG CCCCCTGGACTTTGCTGTTTG ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 4. Primer sequences for PCR (5’-3’) Gene Forward Reverse HRD1-pDsred CCCAAGCTTATGTTCCGCACCGCAGT GGGGTACCCAGTGGGCAACAGGGG HRD1-pCMV- Flag GGGGTACCATGTTCCGCACCGCAGT CCCAAGCTTGTGGGCAACAGGGGACT C HRD1-pCMV-HA GGCCATGGGCCATATGGGATCCTTCC AGGGATGCCACCCGGGGATCCTCAGT GCACCGCAGTGATG GGGCAACAGGGGAC HRD1-N-HA GGCCATGGGCCATATGGGATCCTTCC