Fibroblastic Reticular Cells Predict Response to Immune Checkpoint Inhibitors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fine Tuning by Human Cd1e of Lipid-Specific Immune Responses
Fine tuning by human CD1e of lipid-specific immune responses Federica Facciottia, Marco Cavallaria, Catherine Angénieuxb, Luis F. Garcia-Allesc, François Signorino-Gelob, Lena Angmana, Martine Gilleronc, Jacques Prandic, Germain Puzoc, Luigi Panzad, Chengfeng Xiae, Peng George Wangf, Paolo Dellabonag, Giulia Casoratig, Steven A. Porcellih, Henri de la Salleb, Lucia Moria,i,1, and Gennaro De Liberoa,1 aExperimental Immunology, Department of Biomedicine, University Hospital Basel, 4031 Basel, Switzerland; bInstitut National de la Santé et de la Recherche Médicale, Unité Mixte de Recherche S725, Biology of Human Dendritic Cells, Université de Strasbourg and Etablissement Français du Sang-Alsace, 67065 Strasbourg, France; cCentre National de la Recherche Scientifique, Institut de Pharmacologie et de Biologie Structurale, 31077 Toulouse, France; dDepartment of Chemistry, Food, Pharmaceuticals, and Pharmacology, Università del Piemonte Orientale, 28100 Novara, Italy; eState Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650204, China; fDepartment of Biochemistry and Chemistry, The Ohio State University, Columbus, OH 43210; gExperimental Immunology Unit, Division of Immunology, Transplantation, and Infectious Diseases, Dipartimento di Biotecnologie (DIBIT), San Raffaele Scientific Institute, 20132 Milano, Italy; hAlbert Einstein College of Medicine, Bronx, NY 10461; and iSingapore Immunology Network, Agency for Science Technology and Research, Biopolis, Singapore 138648 Edited by Peter Cresswell, Yale University School of Medicine, New Haven, CT, and approved July 20, 2011 (received for review June 5, 2011) CD1e is a member of the CD1 family that participates in lipid an- molecules traffic to early recycling endosomes and, to a lesser tigen presentation without interacting with the T-cell receptor. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Tools for Cell Therapy and Immunoregulation
RnDSy-lu-2945 Tools for Cell Therapy and Immunoregulation Target Cell TIM-4 SLAM/CD150 BTNL8 PD-L2/B7-DC B7-H1/PD-L1 (Human) Unknown PD-1 B7-1/CD80 TIM-1 SLAM/CD150 Receptor TIM Family SLAM Family Butyrophilins B7/CD28 Families T Cell Multiple Co-Signaling Molecules Co-stimulatory Co-inhibitory Ig Superfamily Regulate T Cell Activation Target Cell T Cell Target Cell T Cell B7-1/CD80 B7-H1/PD-L1 T cell activation requires two signals: 1) recognition of the antigenic peptide/ B7-1/CD80 B7-2/CD86 CTLA-4 major histocompatibility complex (MHC) by the T cell receptor (TCR) and 2) CD28 antigen-independent co-stimulation induced by interactions between B7-2/CD86 B7-H1/PD-L1 B7-1/CD80 co-signaling molecules expressed on target cells, such as antigen-presenting PD-L2/B7-DC PD-1 ICOS cells (APCs), and their T cell-expressed receptors. Engagement of the TCR in B7-H2/ICOS L 2Ig B7-H3 (Mouse) the absence of this second co-stimulatory signal typically results in T cell B7-H1/PD-L1 B7/CD28 Families 4Ig B7-H3 (Human) anergy or apoptosis. In addition, T cell activation can be negatively regulated Unknown Receptors by co-inhibitory molecules present on APCs. Therefore, integration of the 2Ig B7-H3 Unknown B7-H4 (Mouse) Receptors signals transduced by co-stimulatory and co-inhibitory molecules following TCR B7-H5 4Ig B7-H3 engagement directs the outcome and magnitude of a T cell response Unknown Ligand (Human) B7-H5 including the enhancement or suppression of T cell proliferation, B7-H7 Unknown Receptor differentiation, and/or cytokine secretion. -

Dendritic Cell Subsets in Intestinal Immunity and Inflammation Tian Sun, Albert Nguyen and Jennifer L
Dendritic Cell Subsets in Intestinal Immunity and Inflammation Tian Sun, Albert Nguyen and Jennifer L. Gommerman This information is current as J Immunol 2020; 204:1075-1083; ; of September 28, 2021. doi: 10.4049/jimmunol.1900710 http://www.jimmunol.org/content/204/5/1075 Downloaded from References This article cites 152 articles, 56 of which you can access for free at: http://www.jimmunol.org/content/204/5/1075.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on September 28, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2020 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Dendritic Cell Subsets in Intestinal Immunity and Inflammation Tian Sun, Albert Nguyen, and Jennifer L. Gommerman The mammalian intestine is a complex environment shaping the ensuing immune response. In this Brief Review,we that is constantly exposed to Ags derived from food, review the different types of classical DC (cDC) located in the microbiota, and metabolites. -

CD200, Human Recombinant Recombinant Human CD200, : 408-493-1800 | Fax: 408-493-1801 408-493-1801 Fax: | 408-493-1800
BioVision 1/14 For research use only CD200, human recombinant CATALOG #: 7309-50 50 µg ALTERNATE NAMES: CD200 molecule, MOX1, MOX2, MRC, OX-2 SOURCE: E. coli PURITY: > 90% by SDS-PAGE MOL. WEIGHT: 24.8 kDa (225 aa, 31-232 aa + His tag), confirmed by MALDI-TOF. ENDOTOXIN LEVEL: < 1.0 EU per 1 µg of protein FORM: Liquid FORMULATION: 1 mg/ml in 20 mM Tris-HCl buffer (pH 8.0) containing 0.4M Urea. CD200, human recombinant STORAGE CONDITIONS: Can be stored at 4°C short term (1-2 weeks). For long term storage, aliquot and store at -20°C or - 70°C. Avoid repeated freezing and thawing RELATED PRODUCTS: cycles. • CD1E, human recombinant (Cat. No. 7308-100) • CD226, human recombinant (Cat. No. 7310-100) DESCRIPTION : CD200 is a type-1 membrane glycoprotein, which contains two • CD274, mouse recombinant (Cat. No. 7311-100) immunoglobulin domains, and thus belongs to the immunoglobulin superfamily. Studies of • CD300C, human recombinant (Cat. No. 7312-100) the related genes in mouse and rat suggest that this gene may regulate myeloid cell • CD3G, human recombinant (Cat. No. 7313-100) activity and delivers an inhibitory signal for the macrophage lineage in diverse tissues. • CD46, human recombinant (Cat. No. 7314-100) Multiple alternatively spliced transcript variants that encode different isoforms have been • CD5, human recombinant (Cat. No. 7315-100) • CD7, human recombinant (Cat. No. 7316-100) found for this gene. Recombinant human CD200 protein, fused to His-tag at N-terminus, • CD74, human recombinant (Cat. No. 7317-100) was expressed in E.coli. • CD79B, human recombinant (Cat. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Dendritic Cell Progenitor Trafficking and Identification and Functional Analyses of Dendritic Cells with Distinct Developmental Origin
Dissertation zum Erwerb des Doctor of Philosophy (Ph.D.) an der Medizinischen Fakultät der Ludwig-Maximilians-Universität zu München Dendritic cell progenitor trafficking and identification and functional analyses of dendritic cells with distinct developmental origin vorgelegt von: Johanna Marie Salvermoser aus: Pfaffenhofen an der Ilm Jahr: 2020 _____________________ First supervisor: Prof. Dr. Barbara Schraml Second supervisor: Prof. Dr. Anne Krug Third supervisor: Prof. Dr. Christian Schulz Fourth supervisor: PD. Dr. rer. nat. Caspar Ohnmacht Dean: Prof. Dr. med. dent. Reinhard Hickel Datum der Verteidigung: 09.03.2020 2 Abstract ABSTRACT Conventional dendritic cells (cDCs), are the major antigen-presenting cell type that bridges the innate and adaptive immune system. DCs are constantly replenished from myeloid bone marrow progenitors which latest stage, pre-cDC, leave the BM, seeds the peripheral tissues and further differentiates into two functionally and developmentally distinct subsets, cDC1 and cDC2. This study aimed to investigate DC development by assessing the trafficking of pre-cDCs and by analyzing the effect of a specific depletion of DC progenitors. The signals that regulate the recruitment of pre-cDCs to different peripheral organs are poorly understood. Therefore, this study aimed to identify pre-cDCs in different peripheral organs and to find differences in expression pattern of trafficking receptors. In this study 39 trafficking receptors have been identified to be expressed on pre-cDCs of the analysed tissues and showed differences in the expression patterns between peripheral organs. These receptors are interesting candidates to further study differences in the recruitment of pre-cDCs to different peripheral tissues This can provide possibilities to influence the recruitment of pre-cDCs in certain diseases, where the replenishment of cDCs is accelerated. -

Targeting Dendritic Cells with Antigen-Delivering Antibodies for Amelioration of Autoimmunity in Animal Models of Multiple Sclerosis and Other Autoimmune Diseases
antibodies Review Targeting Dendritic Cells with Antigen-Delivering Antibodies for Amelioration of Autoimmunity in Animal Models of Multiple Sclerosis and Other Autoimmune Diseases Courtney A. Iberg and Daniel Hawiger * Department of Molecular Microbiology and Immunology, Saint Louis University School of Medicine, Doisy Research Center, 1205 Carr Lane, St. Louis, MO 63104, USA; [email protected] * Correspondence: [email protected] Received: 31 March 2020; Accepted: 30 April 2020; Published: 15 June 2020 Abstract: The specific targeting of dendritic cells (DCs) using antigen-delivering antibodies has been established to be a highly efficient protocol for the induction of tolerance and protection from autoimmune processes in experimental autoimmune encephalomyelitis (EAE), a model of multiple sclerosis (MS), as well as in some other animal disease models. As the specific mechanisms of such induced tolerance are being investigated, the newly gained insights may also possibly help to design effective treatments for patients. Here we review approaches applied for the amelioration of autoimmunity in animal models based on antibody-mediated targeting of self-antigens to DCs. Further, we discuss relevant mechanisms of immunological tolerance that underlie such approaches, and we also offer some future perspectives for the application of similar methods in certain related disease settings such as transplantation. Keywords: dendritic cells; tolerance; antigen targeting; chimeric antibodies; autoimmunity; multiple sclerosis; diabetes 1. Introduction Over one hundred years ago, Paul Ehrlich coined the term “horror autotoxicus” to define an immune attack against an organism’s healthy tissues [1]. Since then, our knowledge of the complex mechanisms of the immune system as well as our understanding of the pathogenesis of specific autoimmune diseases have grown tremendously. -

A Single Gene Expressed in Fibroblastic Reticular Cells Predicts
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.19.955666; this version posted February 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. A single gene expressed in fibroblastic reticular cells predicts response to cancer immunotherapy Daniele Biasci1,2, , James Thaventhiran1,2*, and Simon Tavaré2,3,* 1MRC Toxicology Unit, University of Cambridge, Robinson Way, Cambridge, CB2 0RE, United Kingdom 2Cancer Research UK Cambridge Institute, Robinson Way, Cambridge CB2 0RE, United Kingdom 3Herbert and Florence Irving Institute for Cancer Dynamics, Columbia University, Schermerhorn Hall, Suite 601, 1190 Amsterdam Ave, New York, NY 10027, United States 1 While the role of CD8+ T cells in mediating response to cancer 46 CD19, FCER2 (CD23), PAX5, BANK1, VPREB3, TCL1A, 2 immunotherapy is well established, the role of other cell types, 47 CLEC17A and FDCSP (Fig. 1A and Table S1). Literature 3 including B cells, remains more controversial. By conducting a 48 reports that these genes are predominantly expressed in B 4 large gene expression study of response to immune checkpoint 49 cells (12), with the exception of FDCSP, which was found to 5 blockade (ICB), here we show that pre-treatment expression of 50 be expressed follicular dendritic cells (FDCs) isolated from 6 B cell genes is associated with ICB response independently of 51 secondary lymphoid organs, but not in B cells (13, 14). In 7 CD8+ T cells. -

Dux4r, Znf384r and PAX5-P80R Mutated B-Cell Precursor Acute
Acute Lymphoblastic Leukemia SUPPLEMENTARY APPENDIX DUX4r , ZNF384r and PAX5 -P80R mutated B-cell precursor acute lymphoblastic leukemia frequently undergo monocytic switch Michaela Novakova, 1,2,3* Marketa Zaliova, 1,2,3* Karel Fiser, 1,2* Barbora Vakrmanova, 1,2 Lucie Slamova, 1,2,3 Alena Musilova, 1,2 Monika Brüggemann, 4 Matthias Ritgen, 4 Eva Fronkova, 1,2,3 Tomas Kalina, 1,2,3 Jan Stary, 2,3 Lucie Winkowska, 1,2 Peter Svec, 5 Alexandra Kolenova, 5 Jan Stuchly, 1,2 Jan Zuna, 1,2,3 Jan Trka, 1,2,3 Ondrej Hrusak 1,2,3# and Ester Mejstrikova 1,2,3# 1CLIP - Childhood Leukemia Investigation Praguerague, Czech Republic; 2Department of Paediatric Hematology and Oncology, Second Faculty of Medicine, Charles University, Prague, Czech Republic; 3University Hospital Motol, Prague, Czech Republic; 4Department of In - ternal Medicine II, University Hospital Schleswig-Holstein, Kiel, Germany and 5Comenius University, National Institute of Children’s Dis - eases, Bratislava, Slovakia *MN, MZ and KF contributed equally as co-first authors. #OH and EM contributed equally as co-senior authors. ©2021 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2020.250423 Received: February 18, 2020. Accepted: June 25, 2020. Pre-published: July 9, 2020. Correspondence: ESTER MEJSTRIKOVA - [email protected] Table S1. S1a. List of antibodies used for diagnostic immunophenotyping. Antibody Fluorochrome Clone Catalogue number Manufacturer CD2 PE 39C1.5 A07744 Beckman Coulter CD3 FITC UCHT1 1F-202-T100 Exbio CD4 PE-Cy7 -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -
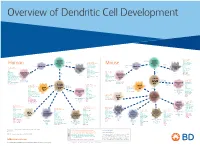
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit)