Identification of Co-Occurring Branchinecta Fairy Shrimp Species from Encysted Embryos Using Multiplex Polymerase Chain Reaction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Fig. Ap. 2.1. Denton Tending His Fairy Shrimp Collection
Fig. Ap. 2.1. Denton tending his fairy shrimp collection. 176 Appendix 1 Hatching and Rearing Back in the bowels of this book we noted that However, salts may leach from soils to ultimately if one takes dry soil samples from a pool basin, make the water salty, a situation which commonly preferably at its deepest point, one can then "just turns off hatching. Tap water is usually unsatis- add water and stir". In a day or two nauplii ap- factory, either because it has high TDS, or because pear if their cysts are present. O.K., so they won't it contains chlorine or chloramine, disinfectants always appear, but you get the idea. which may inhibit hatching or kill emerging If your desire is to hatch and rear fairy nauplii. shrimps the hi-tech way, you should get some As you have read time and again in Chapter 5, guidance from Brendonck et al. (1990) and temperature is an important environmental cue for Maeda-Martinez et al. (1995c). If you merely coaxing larvae from their dormant state. You can want to see what an anostracan is like, buy some guess what temperatures might need to be ap- Artemia cysts at the local aquarium shop and fol- proximated given the sample's origin. Try incu- low directions on the container. Should you wish bation at about 3-5°C if it came from the moun- to find out what's in your favorite pool, or gather tains or high desert. If from California grass- together sufficient animals for a study of behavior lands, 10° is a good level at which to start. -

Vernal Pool Fairy Shrimp (Branchinecta Lynchi)
Invertebrates Vernal Pool Fairy Shrimp (Branchinecta lynchi) Vernal Pool Fairy Shrimp (Brachinecta lynchi) Status State: Meets the requirements as a “rare, threatened, or endangered species” under CEQA Federal: Threatened Critical Habitat: Designated 2006 (USFWS 2006) Population Trend Global: Declining due to habitat loss and fragmentation (Eriksen and Belk 1999) State: As above Within Inventory Area: Unknown Data Characterization The location database for the vernal pool fairy shrimp (Brachinecta lynchi) within the inventory area includes 6 records from 1993, 1997, and 1999. The majority of locations are vernal pools within non-native grassland. Other natural and artificial habitats have a high probability of being occupied by additional populations of the vernal pool fairy shrimp throughout the grassland habitats within the ECCC HCP/NCCP inventory area. Beyond the original description (Eng et al. 1990), a scanning electron micrograph of the cyst (resting egg) (Hill and Shepard 1997), and some generalized natural history data (Helm 1997), no peer-reviewed technical literature has been published concerning the vernal pool fairy shrimp. Eriksen and Belk (1999) presented a brief discussion of the vernal pool fairy shrimp and provided a distribution map. Range The vernal pool fairy shrimp is found from Jackson County near Medford, Oregon, throughout the Central Valley, and west to the central Coast Ranges. Isolated southern populations occur on the Santa Rosa Plateau and near Rancho California in Riverside County (Eng et al.1990, Eriksen -

California Flats Solar Project
Appendix E.10 Dry Season Sampling DRY SEASON SAMPLING, INCLUDING GENETIC ANALYSIS OF CYSTS FOR FEDERALLY LISTED LARGE BRANCHIOPODS AT THE CALIFORNIA FLATS SOLAR PROJECT Prepared for: H.T. HARVEY & ASSOCIATES 983 University Avenue, Building D Los Gatos, CA 95032 Contact: Kelly Hardwicke (408) 458-3236 Prepared by: HELM BIOLOGICAL CONSULTING 4600 Karchner Road Sheridan, CA 95681 Contact: Brent Helm (530) 633-0220 September 2013 DRY SEASON SAMPLING, INCLUDING GENETIC ANALYSIS OF CYSTS FOR FEDERALLY LISTED LARGE BRANCHIOPODS AT THE CALIFORNIA FLATS SOLAR PROJECT INTRODUCTION Helm Biological Consulting (HBC) was contracted by H. T. Harvey & Associates (HTH) to conduct dry-season sampling for the presence of large branchiopods (fairy shrimp, tadpole shrimp, and clam shrimp) that are listed as threatened or endangered under the federal Endangered Species Act (e.g., vernal pool fairy shrimp [Branchinecta lynchi] and longhorn fairy shrimp [Branchinecta longiantenna]) at the California Flats Solar Project (aka “Project”). The contract also included the genetic analysis of any fairy shrimp or tadpole shrimp cysts (embryonic eggs) observed to determine species. The proposed Project consists of approximately 2,615.3 acres (ac) and includes the construction and operation of a 280-megawatt alternating current photovoltaic solar power facility. However, surveys for listed large branchiopods were conducted on a much larger area (i.e., 4,653.45-ac) designated as the “Biological Study Area” (BSA) for the Project (Figure 1), as per direction from U.S. Fish and Wildlife Service (USFWS, Douglas pers. comm. 2011). The BSA is located in southeastern unincorporated Monterey County, California, with an access road to Highway 41 that extends south into northern San Luis Obispo County (Figure 1). -

Longhorn Fairy Shrimp (Branchinecta Longiantenna)
species is known from the Flying M Ranch, on University of California lands, and on the Ichord Ranch where it is currently threatened by indirect and cumulative effects associated with the development of the University of California, Merced campus. e. Conservation Efforts On September 19, 1994, the final rule to list the Conservancy fairy shrimp as endangered was published in the Federal Register (U.S. Fish and Wildlife Service 1994a). In 2005, critical habitat was designated for the Conservancy fairy shrimp and several other vernal pool species in Final Designation of Critical Habitat for Four Vernal Pool Crustaceans and Eleven Vernal Pool Plants in California and Southern Oregon; Evaluation of Economic Exclusions From August 2003 Final Designation; Final Rule (U.S. Fish and Wildlife Service 2005). Within the Northeastern Sacramento Vernal Pool Region, the Conservancy fairy shrimp is protected at the Vina Plains preserve owned by the Nature Conservancy. In the Solano-Colusa Vernal Pool Region the Conservancy fairy shrimp is protected on lands within the Jepson Prairie Ecosystem, including the Burke Ranch and the Jepson Prairie Preserve owned by the Solano County Open Space and Farmland Conservancy and jointly managed by the University of California Reserve System at the Sacramento National Wildlife Refuge in Glenn County and in the Tule Ranch unit of the California Department of Fish and Game Yolo Basin Wildlife Area, in Yolo County. In the San Joaquin Vernal Pool Region, Conservancy fairy shrimp populations are protected at Grasslands Ecological Area on State and federally owned lands, and on the Arena Plains National Wildlife Refuge and the San Luis National Wildlife Refuge in Merced County (California Natural Diversity Database 2005). -

Federal Register/Vol. 65, No. 67/Thursday, April 6, 2000
18026 Federal Register / Vol. 65, No. 67 / Thursday, April 6, 2000 / Proposed Rules By the Board, Chairman Morgan, Vice list, delist, or reclassify a species to evaluate the best scientific and Chairman Burkes, and Commissioner presents substantial information commercial information available as of Clyburn. indicating that the petitioned action that date. The data and information Vernon A. Williams, may be warranted. To the maximum evaluated were to include relevant Secretary. extent practicable, this finding is to be geographic information on the location [FR Doc. 00±8374 Filed 4±5±00; 8:45 am] made within 90 days of the receipt of of vernal pools and fairy shrimp, BILLING CODE 4915±00±P the petition, and the finding is to be including information generated in published promptly in the Federal section 7 consultations since February Register. If the finding is that 29, 1996. DEPARTMENT OF THE INTERIOR substantial information was presented, On September 19, 1994, we published we will commence a status review of the the final rule to list the vernal pool fairy Fish and Wildlife Service involved species. shrimp and vernal pool tadpole shrimp On February 29, 1996, we received a as threatened and endangered, 50 CFR Part 17 petition, dated the same day, to delist respectively, in the Federal Register (59 the vernal pool fairy shrimp FR 48136). The vernal pool fairy shrimp Endangered and Threatened Wildlife (Branchinecta lynchi) and the vernal and vernal pool tadpole shrimp are and Plants; 90-Day Finding on a pool tadpole shrimp (Lepidurus crustacean species endemic to vernal Petition To Delist the Vernal Pool Fairy packardi). -

Conservancy Fairy Shrimp (Branchinecta Conservatio)
2. CONSERVANCY FAIRY SHRIMP (BRANCHINECTA CONSERVATIO) a. Description and Taxonomy Taxonomy.—The Conservancy fairy shrimp (Branchinecta conservatio) was described by Eng, Belk, and Eriksen (Eng et al. 1990). The type specimens were collected in 1982 at Olcott Lake, Solano County, California. The species name was chosen to honor The Nature Conservancy, an organization responsible for protecting and managing a number of vernal pool ecosystems in California, including several that support populations of this species. Description and Identification.—Conservancy fairy shrimp look similar to other fairy shrimp species (Box 1- Appearance and Identification of Vernal Pool Crustaceans). Conservancy fairy shrimp are characterized by the distal segment of the male’s second antennae, which is about 30 percent shorter than the basal segment, and its tip is bent medially about 90 degrees (Eng et al. 1990). The female brood pouch is fusiform (tapered at each end), typically extends to abdominal segment eight, and has a terminal opening (Eng et al. 1990). Males may be from 14 to 27 millimeters (0.6 to 1.1 inch) in length, and females have been measured between 14.5 and 23 millimeters (0.6 and 0.9 inch) long. Conservancy fairy shrimp can be distinguished from the similar looking midvalley fairy shrimp (Branchinecta mesovallensis) by the shape of two humps on the distal segment of the male's second antennae (Belk and Fugate 2000). The midvalley fairy shrimp's antennae is bent such that the larger of the two humps is anterior (towards the head), whereas this same hump in the Conservancy fairy shrimp is posterior (towards the tail). -

90-Day Protocol Survey Report for U.S. Fish and Wildlife Service Listed Vernal Pool Branchiopods, Thomas Juhasz, May 2010
90-Day Protocol Survey Report For U.S. Fish And Wildlife Service Listed Vernal Pool Branchiopods, Thomas Juhasz, May 2010 90-DAY PROTOCOL SURVEY REPORT FOR U.S. FISH AND WILDLIFE SERVICE LISTED VERNAL POOL BRANCHIOPODS City of Santa Clarita Santa Clarita Valley, Los Angeles County, California July 1, 2011 Prepared for: Impact Sciences Inc. 234 East Colorado Blvd. Suite 205 Pasadena, CA 91101 Prepared by: Thomas Juhasz P.O. Box 90036 Pasadena, California 91109 Phone (626) 755-5123 July 1, 2011 Daryl Koutnik Impact Sciences Inc. 234 East Colorado Blvd. Pasadena, CA 91101 Subject: 90-Day Protocol Survey Report for U.S. Fish and Wildlife Service Listed Vernal Pool Branchiopods Santa Clarita Vernal Pool Project Santa Clarita Valley, Los Angeles County, California Dear Mr. Koutnik: I have performed a Wet-Season Protocol Survey for U.S. Fish and Wildlife Service Listed Vernal Pool Branchiopods of the above referenced property (i.e., “the Site”). This report includes my findings, conclusions, recommendations, and supporting documentation. I appreciate the opportunity to be of service on this project. If you should have any questions regarding this report, or if I can be of further assistance, please contact me at (626) 755-5123. Sincerely, Thomas Juhasz cc: Ms. Julie Vanderwier, Ventura office, USFWS Mr. Chris Dellith, Ventura office, USFWS TABLE OF CONTENTS 1.0 INTRODUCTION ................................................................................................... 1 2.0 SITE DESCRIPTION ............................................................................................. -

How Ecology and Evolution Shape Species Distributions and Ecological Interactions Across Time and Space
HOW ECOLOGY AND EVOLUTION SHAPE SPECIES DISTRIBUTIONS AND ECOLOGICAL INTERACTIONS ACROSS TIME AND SPACE by IULIAN GHERGHEL Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Advisor: Ryan A. Martin Department of Biology CASE WESTERN RESERVE UNIVERSITY January, 2021 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the dissertation of Iulian Gherghel Candidate for the degree of Doctor of Philosophy* Committee Chair Dr. Ryan A. Martin Committee Member Dr. Sarah E. Diamond Committee Member Dr. Jean H. Burns Committee Member Dr. Darin A. Croft Committee Member Dr. Viorel D. Popescu Date of Defense November 17, 2020 * We also certify that written approval has been obtained for any proprietary material contained therein TABLE OF CONTENTS List of tables ........................................................................................................................ v List of figures ..................................................................................................................... vi Acknowledgements .......................................................................................................... viii Abstract ............................................................................................................................. iix INTRODUCTION............................................................................................................. 1 CHAPTER 1. POSTGLACIAL RECOLONIZATION OF NORTH AMERICA BY SPADEFOOT TOADS: INTEGRATING -
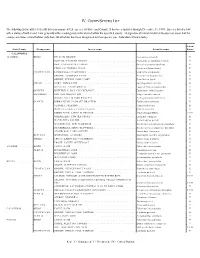
Iv. County/Species List
IV. COUNTY/SPECIES LIST The following list identifies federally listed or proposed U.S. species by State and County. It has been updated through December 31, 1999. Species listed below with a status of both E and T are generally either endangered or threatened within the specified county. Designation of critical habitat (CH) does not mean that the county constitutes critical habitat, only that critical habitat has been designated for that species (see Addendum A Instructions). Action/ State/County Group name Inverse name Scientific name Status CALIFORNIA ALAMEDA ......... BIRDS ....... PELICAN, BROWN ........................................ Pelicanus occidentalis E PLOVER, WESTERN SNOWY ................................ Charadrius alexandrinus nivosus T RAIL, CALIFORNIA CLAPPER ............................... Rallus longirostris obsoletus E TERN, CALIFORNIA LEAST ................................. Sterna antillarum browni E CRUSTACEAN . LINDERIELLA, CALIFORNIA ................................ Linderiella occidentalis E SHRIMP, LONGHORN FAIRY ................................ Branchinecta longiantenna E SHRIMP, VERNAL POOL FAIRY .............................. Branchinecta lynchi T FISHES ...... GOBY, TIDEWATER ...................................... Eucyclogobius newberryi E SPLITTAIL, SACRAMENTO ................................. Pogonichthys macrolepidotus T INSECTS ..... BUTTERFLY, BAY CHECKERSPOT ........................... Euphydryas editha bayensis T MAMMALS ... FOX, SAN JOAQUIN KIT .................................... Vulpes macrotis -

VERNAL POOL FAIRY SHRIMP Branchinecta Lynchi
U.S. Fish & Wildlife Service Sacramento Fish & Wildlife Office Species Account VERNAL POOL FAIRY SHRIMP Branchinecta lynchi CLASSIFICATION: Threatened Federal Register 59-48136; September 19, 1994 http://ecos.fws.gov/docs/federal_register/fr2692.pdf On October 9, 2007, we published a 5-year review recommending that the species remain listed as threatened. CRITICAL HABITAT: Designated Originally designated in Federal Register 68:46683; August 6, 2003. The designation was revised in FR 70:46923; August 11, 2005. Species by unit designations were published in FR 71:7117 (PDF), February 10, 2006. RECOVERY PLAN: Final Recovery Plan for Vernal Pool Ecosystems of California and Southern Oregon, December 15, 2005. http://ecos.fws.gov/docs/recovery_plan/060614.pdfhttp://ecos.fws.gov/docs/recovery_plan/0606 14.pdf DESCRIPTION The vernal pool fairy shrimp (Branchinecta lynchi), is a small crustacean in the Branchinectidae family. It ranges in size from ½ to one inch long. Fairy shrimp feed on algae, bacteria, protozoa, rotifers and bits of detritus. Fairy shrimp are aquatic species in the order Anostraca. They have delicate elongate bodies, large stalked compound eyes, no carapaces, and eleven pairs of swimming legs. They glide gracefully upside down, swimming by beating their legs in a complex, wavelike movement that passes from front to back. The vernal pool fairy shrimp occupies a variety of different vernal pool habitats, from small, clear, sandstone rock pools to large, turbid, alkaline, grassland valley floor pools. Although the species has been collected from large vernal pools, including one exceeding 25 acres, it tends to occur in smaller pools. It is most frequently found in pools measuring less than 0.05 acre. -

Agate Desert Vernal Pool Final Draft Functional Assessment Methodology
AGATE DESERT VERNAL POOL FINAL DRAFT FUNCTIONAL ASSESSMENT METHODOLOGY Prepared for: April 2007 Agate Desert Technical Advisory Committee In association with Adamus Resource Assessment, Inc. Paul Adamus, Ph.D. AGATE DESERT VERNAL POOL FINAL DRAFT FUNCTIONAL ASSESSMENT METHODOLOGY Prepared for: April 2007 Agate Desert Technical Advisory Committee 8950 Cal Center Drive Building 3, Suite 300 Sacramento, CA 95826 916.564.4500 www.esassoc.com Los Angeles Oakland Petaluma Portland San Diego San Francisco Seattle Tampa Woodland Hills 204081 In association with Adamus Resource Assessment, Inc. Paul Adamus, Ph.D. TABLE OF CONTENTS Agate Desert Vernal Pool Functional Assessment Methodology Page Acknowledgements iii 1 Introduction 1-1 1.1 Unique Ecology of Vernal Pool Wetlands 1-7 1.2 Relationship of Method to Wetland Classification Schemes 1-10 2 Background 2-1 2.1 Overview of Vernal Pools in the Agate Desert 2-1 2.2 Wetland Planning and Conservation in Oregon 2-4 2.3 Agate Desert Technical Advisory Committee 2-5 2.4 Agate Desert Stakeholder Committee 2-5 3 Method Development and Application 3-1 3.1 Indicator and Scoring Notes 3-1 3.2 Method Development 3-2 3.3 Method Application 3-3 3.4 Functions and Values for Vernal Pool Wetland Assessment 3-5 3.5 Functions and Values Selected for Vernal Pools 3-6 3.6 Multiple Scales and Assessment Site Classification 3-18 3.7 Landscape and Pool Indicators 3-19 3.8 Derived Indicators 3-25 3.9 Indicator Scoring Analyses 3-25 4 Scoring Models 4-1 4.1 Scoring Model Development 4-1 4.2 Summary Lists of Indicators -

Western Riverside County Regional Conservation Authority (RCA) Annual Report to the Wildlife Agencies
Western Riverside County Multiple Species Habitat Conservation Plan (MSHCP) Biological Monitoring Program Vernal Pool Survey Report 2010 Riverside Fairy Shrimp pictured 8 April 2011 Vernal Pool Survey Report 2010 TABLE OF CONTENTS INTRODUCTION.........................................................................................................................................3 GOALS AND OBJECTIVES ...........................................................................................................2 RIVERSIDE FAIRY SHRIMP.........................................................................................................2 VERNAL POOL FAIRY SHRIMP .................................................................................................2 WESTERN SPADEFOOT................................................................................................................3 METHODS ....................................................................................................................................................3 PROTOCOL DEVELOPMENT .......................................................................................................3 PERSONNEL AND TRAINING......................................................................................................3 SITE SELECTION ...........................................................................................................................4 SURVEY METHODS ......................................................................................................................4