Transferability of Microsatellite Markers in Syagrus Coronata (Mart.) Becc
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Winter-Fall Sale 2002 Palm Trees-Web
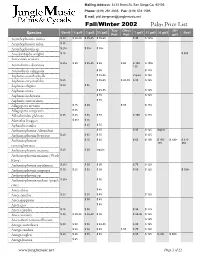
Mailing Address: 3233 Brant St. San Diego Ca, 92103 Phone: (619) 291 4605 Fax: (619) 574 1595 E mail: [email protected] Fall/Winter 2002 Palm Price List Tree Citrus 25/+ Band$ 1 gal$ 2 gal$ 3/5 gal$ 7 gal$ 15 gal$ 20 gal$ Box$ Species Pot$ Pot$ gal$ Acanthophoenix crinita $ 30 $ 30-40 $ 35-45 $ 55-65 $ 95 $ 125+ Acanthophoenix rubra $ 35 Acanthophoenix sp. $ 25+ $ 35+ $ 55+ Acoelorrhaphe wrightii $ 15 $ 300 Acrocomia aculeata $ 25+ $ 35 $ 35-45 $ 65 $ 65 $ 100- $ 150+ Actinokentia divaricata 135 Actinorhytis calapparia $ 55 $ 125 Aiphanes acanthophylla $ 45-55 inquire $ 125 Aiphanes caryotaefolia $ 25 $ 55-65 $ 45-55 $ 85 $ 125 Aiphanes elegans $ 20 $ 35 Aiphanes erosa $ 45-55 $ 125 Aiphanes lindeniana $ 55 $ 125 Aiphanes vincentsiana $ 55 Allagoptera arenaria $ 25 $ 40 $ 55 $ 135 Allagoptera campestris $ 35 Alloschmidtia glabrata $ 35 $ 45 $ 55 $ 85 $ 150 $ 175 Alsmithia longipes $ 35+ $ 55 Aphandra natalia $ 35 $ 55 Archontophoenix Alexandrae $ 55 $ 85 $ 125 inquire Archontophoenix Beatricae $ 20 $ 35 $ 55 $ 125 Archontophoenix $ 25 $ 45 $ 65 $ 100 $ 150- $ 200+ $ 310- 175 350 cunninghamiana Archontophoenix maxima $ 25 $ 30 inquire Archontophoenix maxima (Wash River) Archontophoenix myolaensis $ 25+ $ 30 $ 50 $ 75 $ 125 Archontophoenix purpurea $ 30 $ 25 $ 35 $ 50 $ 85 $ 125 $ 300+ Archontophoenix sp. Archontophoenix tuckerii (peach $ 25+ $ 55 river) Areca alicae $ 45 Areca catechu $ 20 $ 35 $ 45 $ 125 Areca guppyana $ 30 $ 45 Areca ipot $ 45 Areca triandra $ 25 $ 30 $ 95 $ 125 Areca vestiaria $ 25 $ 30-35 $ 35-40 $ 55 $ 85-95 $ 125 Arecastrum romanzoffianum $ 125 Arenga australasica $ 20 $ 30 $ 35 $ 45-55 $ 85 $ 125 Arenga caudata $ 20 $ 30 $ 45 $ 55 $ 75 $ 100 Arenga engleri $ 20 $ 60 $ 35 $ 45 $ 85 $ 125 $ 200 $ 300+ Arenga hastata $ 25 www.junglemusic.net Page 1 of 22 Tree Citrus 25/+ Band$ 1 gal$ 2 gal$ 3/5 gal$ 7 gal$ 15 gal$ 20 gal$ Box$ Species Pot$ Pot$ gal$ Arenga hookeriana inquire Arenga micranthe 'Lhutan' $ 20 inquire Arenga pinnata $ 35 $ 50 $ 85 $ 125 Arenga sp. -

Essential Oils and Oil from Seeds of Syagrus Coronata
Vol. 10(23), pp. 310-317, 17 June, 2016 DOI: 10.5897/JMPR2016.6098 Article Number: 89CD55858997 ISSN 1996-0875 Journal of Medicinal Plants Research Copyright © 2016 Author(s) retain the copyright of this article http://www.academicjournals.org/JMPR Full Length Research Paper Syagrus coronata seed oils have antimicrobial action against multidrug-resistant Staphylococcus aureus Cibele Maria Alves da Silva Bessa1, Rodrigo Santana do Nascimento1, Renata Carla Corrêa Alves1*, José Matias Anselmo2, Ana Paula Sant'Anna da Silva1, Alexandre Gomes da Silva1, Vera Lúcia de Menezes Lima1, Josean Fechine Tavares3, Luís Cláudio Nascimento da Silva1,2, Márcia Vanusa da Silva1 and Maria Tereza dos Santos Correia1 1Departamento de Bioquímica, Centro de Ciências Biológicas, Universidade Federal de Pernambuco, Av. Professor Moraes Rego, Cidade Universitária, 1235, 50670-901, Recife, Pernambuco, Brazil. 2Faculdade Pernambucana de Saúde, Av. Jean Emile Favre, 420, Imbiribeira, 51200-060, Recife, Pernambuco, Brazil. 3Departamento de Ciências Farmacêuticas, Universidade Federal da Paraíba, Campus I, Castelo Branco, 58051-970, Joao Pessoa, Paraíba, Brazil. Received 15 March, 2016; Accepted 20 May, 2016 Syagrus coronata (Mart.) Becc. (Arecaceae) is a native Brazilian palm (ouricuri) and despite the use of its derived products by traditional communities, few scientific reports have been published regarding its biomedical activity. This study investigates the chemical composition and anti-Staphylococcus aureus effects of both manufactured oil (SCO) and essential oil (SCEO) from S. coronata seeds. SCO was provided by rural inhabitants, while SCEO was obtained by hydrodistillation. Chemical characterization was performed by gas chromatography-mass spectrometry (GC/MS). In vitro antimicrobial activity was determined against 17 S. aureus strains, including multidrug-resistant strains. -

MORPHOLOGY of FRUITS, DIASPORES, SEEDS, SEEDLINGS, and SAPLINGS of Syagrus Coronata (Mart.) Becc
652 Original Article MORPHOLOGY OF FRUITS, DIASPORES, SEEDS, SEEDLINGS, AND SAPLINGS OF Syagrus coronata (Mart.) Becc. MORFOLOGIA DE FRUTOS, DIÁSPOROS, SEMENTES, PLÂNTULAS E MUDAS DE Syagrus coronata (Mart.) Becc Sueli da Silva SANTOS-MOURA 1; Edilma Pereira GONÇALVES 2; Luan Danilo Ferreira de Andrade MELO 1; Larissa Guimarães PAIVA 1; Tatiana Maria da SILVA 1 1. Master's in Agricultural Production by the Rural Federal University of Pernambuco, Academic Unit of Garanhuns, Garanhuns, PE, Brazil; 2. Teacher, doctor at the Federal Rural University of Pernambuco, Academic Unit of Garanhuns, Garanhuns, PE, Brazil. ABSTRACT: Licuri ( Syagrus coronata (Mart.) Becc.) is an ornamental palm tree native of Brazil with great economic potential, because it provides raw material for manufacturing a wide range of products. The objective of this study was to assess the morphology of the fruits, diaspores, seeds, seedlings, and saplings of Syagrus coronata . The study was performed at the Laboratory of Seed Analysis (LSA) of the Federal Rural University of Pernambuco/Academic Unit of Garanhuns-PE, by using licuri fruits collected from the rural area of Caetés-PE. It was evaluated fruit morphology, diaspores, seeds, seedlings and saplings. Germination, in the form of cotyledon petiole emergence, began 15 days after sowing, is hypogeal, cryptocotylar, and remote tubular. It is slow and uneven, extending up to 60 days after the first eophyll appears. The saplings have alternate, pinnate, glabrous, entire leaves with parallel venation and sheath invagination. The primary roots persistent, the secondary roots arise from the stem root node in the primary root, and lateral roots only fasciculate was evidenced when the change was 300 days, and must remain in the nursery for at least 360 days after germination before taking it to the field, due to the slow development of this species. -

Sfps Fall 2011 Sale Plant List
SFPS FALL 2011 SALE PLANT LIST PLANTS VENDOR # Palms Acanthophoenix rubra 35 Acoelorrhaphe wrightii 26, 67 Acrocomia aculeata 50, 67 Actinokentia divaricata 35, 57, 66, 68, 72 Actinorhytis calapparia 72 Adonidia merrillii 31, 57, 66, 89 Adonidia merrillii var. "Golden Form" 35 Aiphanes aculeata = Aiphanes horrida - Aiphanes caryotifolia = Aiphanes horrida - Aiphanes erosa = Aiphanes minima - Aiphanes horrida 35, 68, 72 Aiphanes minima 68 Aiphanes vincentiana = Aiphanes minima - Allagoptera arenaria 57, 66, 67, 68, 72 Allagoptera campestris 67 Allagoptera leucocalyx 57 Alloschmidia glabrata = Basselinia glabrata - Alsmithia longipes = Heterospathe longipes - Archontophoenix cunninghamiana var. 'Illawara' 68 Archontophoenix maxima 67, 72 Archontophoenix myolensis 50, 66, 67, 68 Archontophoenix purpurea 57, 66, 72 Archontophoenix tuckeri 66, 68 Areca aliceae = Areca triandra - Areca camarinensis 57, 68 Areca catechu 57, 67, 72 Areca catechu var. 'Dwarf' 35, 50 Areca hutchinsoniana 68 Areca ipot 67 Areca latiloba = Areca montana - Areca macrocalyx var. 'Red Form' 35, 57, 68 Areca macrocarpa 68 Areca montana 57 Areca triandra 68, 72 Areca vestiaria 25, 35, 57, 67, 68 Areca vestiaria var. 'Orange Form' 25, 57, 67, 72 Areca vestiaria var. 'Maroon Leaf' 35, 57, 67 Areca vestiaria var. 'Red Leaf' 57, 67, 72 Areca sp. 'Yellow Crownshaft' 25 Arenga ambong = Arenga undulatifolia - Arenga brevipes 57 Arenga caudata 66 Arenga engleri 31, 66, 68, 72 Arenga hookeriana 35, 57, 66, 72 Arenga microcarpa 26, 66 Arenga obtusifolia 57, 66 PLANTS VENDOR # Arenga pinnata 50, 57, 66, 67, 68 Arenga porphyrocarpa 66 Arenga tremula 26, 57, 66, 68, 72 Arenga undulatifolia 35, 57, 66, 67 Arenga westerhoutii 68 Asterogyne martiana 57, 68, 72 Astrocaryum acaule 72 Astrocaryum alatum 35, 50, 57, 67 Astrocaryum mexicanum 72 Astrocaryum murumuru 72 Attalea butyracea 57, 67, 72 Attalea cohune 35 Attalea phalerata 50, 91 Attalea rostrata 68 Attalea speciosa 50, 66 Bactris bidentula 72 Bactris gasipaes 67 Bactris gasipaes var. -

Record of Tritrophic Relationship Between Syagrus Coronata (Martius)
doi: 10.12741/ebrasilis.v14.e922 e-ISSN 1983-0572 Creative Commons License v4.0 (CC-BY) Copyright © Author(s) Article Full Open Access Scientific Note Record of tritrophic relationship between Syagrus coronata (Martius) Beccari (Arecaceae), Pachymerus nucleorum Fabricius (Coleoptera: Chrysomelidae: Bruchinae) and Heterospilus sp. (Hymenoptera: Braconidae) in the State of Alagoas, northeastern Brazil Jefferson Duarte de Melo , Suianne Oliveira dos Santos Cajé & Iracilda Maria de Moura Lima Laboratório de Bioecologia de Insetos, Instituto de Ciências Biológicas e da Saúde, Universidade Federal de Alagoas, Maceió, Alagoas, Brazil. EntomoBrasilis 14: e922 (2021) Edited by: Abstract. Some conservation units in Brazil border urban areas, like the Catolé and Fernão Velho William Costa Rodrigues Environmental Protection Area (EPA) in the State of Alagoas. In urban areas, there is the habit of cultivating plants for landscape purposes, and Syagrus coronata (Martius) Beccari (Arecaceae), “Licuri” Article History: or “Ouricuri”, is a palm tree commonly used in ornamentation; a native species from Caatinga and Received: 23.vii.2020 Atlantic Forest biomes widely explored through time. Some insects have part of their development Accepted: 30.xii.2020 associated with plants, and Pachymerus nucleorum Fabricius (Coleoptera: Chrysomelidae: Bruchinae) Published: 06.iii.2021 has a close connection with some Arecaceae. Females usually lay eggs on the surface of fallen fruits and the immatures feed on the seed under the drupe endocarp; the larvae, even protected by the Corresponding author: hard surface could be preyed by skilled parasitoid wasps. Here, the record of a tritrophic relationship Jefferson Duarte de Melo between S. coronata, P. nucleorum, and a wasp of the genus Heterospilus (Hymenoptera: Braconidae) [email protected] in an urbanized region of Alagoas, close to a remnant of Atlantic Forest of the Catolé and Fernão Funding agencies: Velho EPA is communicated. -

Evaluating Insect-Host Interactions As a Driver of Species Divergence in Palm Flower Weevils
ARTICLE https://doi.org/10.1038/s42003-020-01482-3 OPEN Evaluating insect-host interactions as a driver of species divergence in palm flower weevils ✉ Bruno A. S. de Medeiros 1,2 & Brian D. Farrell2 1234567890():,; Plants and their specialized flower visitors provide valuable insights into the evolutionary consequences of species interactions. In particular, antagonistic interactions between insects and plants have often been invoked as a major driver of diversification. Here we use a tropical community of palms and their specialized insect flower visitors to test whether antagonisms lead to higher population divergence. Interactions between palms and the insects visiting their flowers range from brood pollination to florivory and commensalism, with the latter being species that feed on decaying–and presumably undefended–plant tissues. We test the role of insect-host interactions in the early stages of diversification of nine species of beetles sharing host plants and geographical ranges by first delimiting cryptic species and then using models of genetic isolation by environment. The degree to which insect populations are structured by the genetic divergence of plant populations varies. A hierarchical model reveals that this variation is largely uncorrelated with the kind of interaction, showing that antag- onistic interactions are not associated with higher genetic differentiation. Other aspects of host use that affect plant-associated insects regardless of the outcomes of their interactions, such as sensory biases, are likely more general drivers of insect population divergence. 1 Smithsonian Tropical Research Institute, Panama City, Panama. 2 Museum of Comparative Zoology, Department of Organismic & Evolutionary Biology, ✉ Harvard University, Cambridge, MA, USA. email: [email protected] COMMUNICATIONS BIOLOGY | (2020) 3:749 | https://doi.org/10.1038/s42003-020-01482-3 | www.nature.com/commsbio 1 ARTICLE COMMUNICATIONS BIOLOGY | https://doi.org/10.1038/s42003-020-01482-3 nsects comprise about two-thirds of the 1.5 million described from S. -

Characterization of the Complete Plastid Genome of Butia Eriospatha (Arecaceae)
Genetics and Molecular Biology 43, 4, e20200023 (2020) Copyright © 2020, Sociedade Brasileira de Genética. DOI: https://doi.org/10.1590/1678-4685-GMB-2020-0023 Genome Insight Plant Genetics Characterization of the complete plastid genome of Butia eriospatha (Arecaceae) Jeison Willy de Souza Magnabosco1, Hugo Pacheco de Freitas Fraga1, Raquel Santos da Silva1, Marcelo Rogalski2, Emanuel Maltempi de Souza3, Miguel Pedro Guerra4,5 and Leila do Nascimento Vieira1 1Universidade Federal do Paraná, Programa de Pós-graduação em Botânica, Curitiba, Paraná, Brazil. 2Universidade Federal de Viçosa, Programa de Pós-graduação em Fisiologia Vegetal, Viçosa, MG, Brazil. 3Universidade Federal do Paraná, Programa de Pós-graduação em Bioquímica e Biologia Molecular, Curitiba, PR, Brazil. 4Universidade Federal de Santa Catarina, Programa de Pós-graduação em Recursos Genéticos Vegetais, Florianópolis, SC, Brazil. 5Universidade Federal de Santa Catarina, Programa de Pós-Graduação em Ecossistemas Agrícolas e Naturais, Curitibanos, SC, Brazil. Abstract Butia eriospatha is an endemic palm species from the Atlantic Rainforest in Brazil, a biodiversity hotspot. This species is currently listed in the IUCN red list as vulnerable and lacks specific plastid markers for population genetics studies. In addition, the evolutionary relationship within the genus Butia is not yet well resolved. Here, we sequenced and charac- terized the complete plastid genome (plastome) sequence of B. eriospatha. The complete plastome sequence is 154,048 bp in length, with the typical quadripartite structure. This plastome length and genes content is consistent with other six species from tribe Cocoseae. However, the Inverted Repeat (IR) borders show some variation among the ana- lyzed species from this tribe. Species from the Bactridinae (Astrocaryum and Acrocomia) and Elaeidinae (Elaeis) subtribes present the rps19 gene completely duplicated in the IR region. -

Morphological Characterization and Germination of Syagrus Schizophylla (Mart.) Glass
DOI: 10.14295/CS.v10i1.2997 Comunicata Scientiae 10(1): 54-64, 2019 Article e-ISSN: 2177-5133 www.comunicatascientiae.com Morphological characterization and germination of Syagrus schizophylla (Mart.) Glass. (ARECACEAE) Rômulo André Beltrame*, Janie Mendes Jasmim, Henrique Duarte Vieira State University of North Fluminense *Corresponding author, e-mail: [email protected] Abstract The interest in Syagrus schizophylla as an ornamental palm tree and the demand for conservation and preservation of the species led to this research. The objective was to study the physiological characteristics of its germination at different temperatures, as well as the morphological and biometrical characterization of diaspores and seedlings at the initial stages of growth and development. The research was divided into two experiments. In the first one, the aim was to identify the water absorption phases of seeds during germination under five scarification treatments as follows: intact diaspores, scarified diaspores, diaspores with endocarp rupture and intact seeds. In the second experiment, germination was tested at 25, 30 e 25 - 35 ºC; the first germination count, seedling emergence, abnormal seedlings, non-germinated seeds, the emergence curve, the emergence speed index and the mean time of emergence were evaluated. Afterwards, the morphological and biometrical characteristics of diaspores and seedlings were described. The water absorption curve observed under the different scarification treatments showed different water absorption patterns. Emergence percentages were 53, 61 and 47% at 25, 30 and 25 - 35 ºC, respectively. The highest emergence speed index was obtained at 30 ºC. The mean time of emergence was 30 days, approximately, under all the temperatures tested. The diaspores showed a great variability in both shape and size, presenting a globular to ovoid shape with an average length of 2.44 cm and an average width of 1.39 cm. -

(Syagrus Coronata (Mart.) Becc.) Found in the Atlantic Forest of Minas Gerais, Brazil Ciência E Tecnologia De Alimentos, Vol
Ciência e Tecnologia de Alimentos ISSN: 0101-2061 [email protected] Sociedade Brasileira de Ciência e Tecnologia de Alimentos Brasil de PAULA FILHO, Galdino Xavier; Fontenelle BARREIRA, Tibério; da Cruz RODRIGUES, Vívian Cristina; de Morais CARDOSO, Leandro; Stampini Duarte MARTINO, Hércia; PINHEIRO-SANT’ANA, Helena Maria Study of the physical and physicochemical characteristics of fruits of the licuri palm (Syagrus coronata (Mart.) Becc.) found in the Atlantic Forest of Minas Gerais, Brazil Ciência e Tecnologia de Alimentos, vol. 35, núm. 3, julio-septiembre, 2015, pp. 474-480 Sociedade Brasileira de Ciência e Tecnologia de Alimentos Campinas, Brasil Available in: http://www.redalyc.org/articulo.oa?id=395942248012 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative a ISSN 0101-2061 Food Science and Technology DDOI http://dx.doi.org/10.1590/1678-457X.6652 Study of the physical and physicochemical characteristics of fruits of the licuri palm (Syagrus coronata (Mart.) Becc.) found in the Atlantic Forest of Minas Gerais, Brazil Galdino Xavier de PAULA FILHD1*, Tibério Fontenelle BARREIRA1, Vívian Cristina da Cruz RDDRIGUES1, Leandro de Morais CARDDSD2, Hércia Stampini Duarte MARTIND1, Helena Maria PINHEIRD-SANT’ANA1 Abstract The Atlantic Forest has species of native fruits, consumed fresh and processed, which have an important contribution to food sovereignty of families that consume it. This study examined the physical and physicochemical characteristics, proximate composition, concentration of carotenoids, vitamin C, vitamin E and minerals in the pulp and kernels of fruits of licuri (Syagrus coronata (Mart.) Becc.). -

Thaíssa Scalco Caixeta Germinação De Sementes De Palmeira Licuri
UNIVERSIDADE JOSÉ DO ROSÁRIO VELLANO - UNIFENAS THAÍSSA SCALCO CAIXETA GERMINAÇÃO DE SEMENTES DE PALMEIRA LICURI Alfenas-MG 2018 2 THAÍSSA SCALCO CAIXETA GERMINAÇÃO DE SEMENTES DE PALMEIRA LICURI Dissertação apresentada à Universidade José do Rosário Vellano – UNIFENAS como parte das exigências para obtenção do título de Mestre em Sistema de Produção na Agropecuária. Orientador: Prof. Dr. Paulo Roberto Corrêa Landgraf Coorientadora: Profª Drª Patricia de Oliveira Alvim Veiga Alfenas-MG 2018 3 Dados internacionais de catalogação-na-publicação Biblioteca Central da UNIFENAS Caixeta, Thaíssa Scalco Germinação de sementes de palmeira licuri / Thaíssa Scalco Caixeta — Alfenas, 2018. 40 f. o Orientador: Prof. Dr. Paulo Roberto Corrêa Landgraf Co-orientadora: Prof.ª Drª. Patrícia de Oliveira Alvim Veiga Dissertação (Mestrado) - Programa de Pós-graduação em Sistema de Produção na Agropecuária. – Universidade José do Rosário Vellano, Alfenas, 2018. 1. Syagrus Coronata. 2.Substratos. 3.Tempo de Embebição. I. Universidade José do Rosário Vellano II. Título CDU 633.855.34(043.3) Samira Vidal da Silva Ramos Bibliotecária CRB6 3474 4 5 Dedico a todos que, de algum modo, me incentivaram e me acompanharam durante a realização desta dissertação. 6 AGRADECIMENTOS Manifesto minha gratidão, pois a caminhada não é sozinha, mas sim cercada de pessoas importantes em nossas vidas: A Deus, gratidão por tudo que tenho e fiz até hoje. Deus é quem me fortalece, ilumina, capacita, concede sabedoria e proteção. À minha mãe Vanessa Scalco Caixeta, a meu pai Isaltino Franco Caixeta e aos familiares que sempre me apoiam e compartilham com paciência das minhas alegrias e vitórias; ao meu namorado Matheus Robin Caixeta pelo amor, pelo carinho, pelo incentivo; e pela alegria em tê-los em minha vida. -
Guide to the Palms of Northeastern Brazil Lnoblick
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/336364606 Guide to the Palms of Northeastern Brazil LNoblick Book · October 2019 CITATIONS READS 0 48 1 author: Larry Noblick Montgomery Botanical Center 59 PUBLICATIONS 423 CITATIONS SEE PROFILE Some of the authors of this publication are also working on these related projects: “Molecular and morphoanatomical phylogeny of the genus Acrocomia (Arecaceae): a taxonomic study of a group of native palm trees with great socioeconomic and environmental interest” View project All content following this page was uploaded by Larry Noblick on 09 October 2019. The user has requested enhancement of the downloaded file. Guide to the Palms of Northeastern Brazil UNIVERSIDADE ESTADUAL DE FEIRA DE SANTANA Evandro do Nascimento Silva Reitor Amali de Angelis Mussi Vice-reitora Eraldo Medeiros Costa Neto Diretor Valdomiro Santana Editor Zenailda Novais Assistente Editorial CONSELHO EDITORIAL Adeítalo Manoel Pinto Antonio César Ferreira da Silva Antônio Vieira da Andrade Neto Diógenes Oliveira Senna Geciara da Silva Carvalho Gilberto Marcos de Mendonça Santos Jorge Aliomar Barreiros Dantas Marluce Nunes Oliveira Nilo Henrique Neves dos Reis Larry R. Noblick e Guide to Guid to the Palms of Northeastern Brazil Feira de Santana 2019 Copyright © 2019 by Larry R. Noblick Projeto gráfico: Ericson Peres Editoração eletrônica: Ericson Peres Capa: Ericson Peres Revisão de provas: Francisco de Assis Ribeiro dos Santos Normalização bibliográfica: Francisco de Assis Ribeiro dos Santos Revisão textual: Francisco de Assis Ribeiro dos Santos Ficha Catalográfica - Biblioteca Central Julieta Carteado - UEFS G971 Guide to the palms of northeastern Brazil [recurso eletrônico] / Larry R. -

The Complete Plastome of Macaw Palm [Acrocomia Aculeata (Jacq.) Lodd
Planta https://doi.org/10.1007/s00425-018-2841-x ORIGINAL ARTICLE The complete plastome of macaw palm [Acrocomia aculeata (Jacq.) Lodd. ex Mart.] and extensive molecular analyses of the evolution of plastid genes in Arecaceae Amanda de Santana Lopes1 · Túlio Gomes Pacheco1 · Tabea Nimz1 · Leila do Nascimento Vieira2 · Miguel P. Guerra2 · Rubens O. Nodari2 · Emanuel Maltempi de Souza3 · Fábio de Oliveira Pedrosa3 · Marcelo Rogalski1 Received: 1 November 2017 / Accepted: 10 January 2018 © Springer-Verlag GmbH Germany, part of Springer Nature 2018 Abstract Main conclusion The plastome of macaw palm was sequenced allowing analyses of evolution and molecular markers. Additionally, we demonstrated that more than half of plastid protein-coding genes in Arecaceae underwent positive selection. Macaw palm is a native species from tropical and subtropical Americas. It shows high production of oil per hectare reaching up to 70% of oil content in fruits and an interesting plasticity to grow in diferent ecosystems. Its domestication and breeding are still in the beginning, which makes the development of molecular markers essential to assess natural populations and germplasm collections. Therefore, we sequenced and characterized in detail the plastome of macaw palm. A total of 221 SSR loci were identifed in the plastome of macaw palm. Additionally, eight polymorphism hotspots were characterized at level of subfamily and tribe. Moreover, several events of gain and loss of RNA editing sites were found within the subfam- ily Arecoideae. Aiming to uncover evolutionary events in Arecaceae, we also analyzed extensively the evolution of plastid genes. The analyses show that highly divergent genes seem to evolve in a species-specifc manner, suggesting that gene degeneration events may be occurring within Arecaceae at the level of genus or species.