Marker -Trait Associations Known in Rosaceae Crops (Table and References Compiled by Dr
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Canadian Viagra Generic
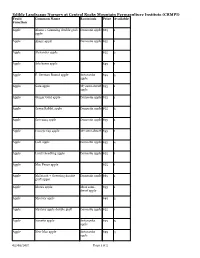
Edible Landscape Nursery at Central Rocky Mountain Permaculture Institute (CRMPI) www.crmpi.org/projects/nursery www.coloradopear.com Additional Fruit/ Container Currently quantities Common Name Rootstock Price Function Size available available summer 2018 Apple Akane + Greening double graft #7 Domestic $65 1 apple apple Apple Alexander apple #10-#15 Domestic $55 1 apple Apple Arkcharm apple #10-#15 Domestic $55 1 apple Apple E. German Rome apple #10-#15 Antonovka $55 3 apple Apple Green Rabbit apple #10-#15 Domestic $55 3 apple Apple Greening apple #10-#15 Domestic $55 1 apple Apple Lodi apple #10-#15 Domestic $55 2 apple Apple Mac Fence apple #10-#15 Domestic $55 1 apple Apple Nanette apple #6-#7 Antonovka $55 3 apple Apple New Mac apple / Novamac #10-#15 Domestic $55 2 apple Apple Northern Spy apple #10-#15 M111 semi- $75 1 dwarf apple Apple Red Baron apple #10-#15 Domestic $55 1 apple Apple Red Gravenstein apple #10-#15 M111 semi- $75 5 dwarf apple Apple Sweet Sixteen apple #10-#15 Antonovka $55 2 apple Apple Yellow transparent apple #10-#15 Antonovka $55 1 apple Apple Wolf River + Gold Rush apple #10-#15 Antonovka $75 1 double graft apple Antique apple Colorado Orange + Gold Rush #10-#15 Antonovka $65 1 double graft apple apple Antique apple Colorado Orange heritage apple #10-#15 Antonovka $55 1 apple Antique apple Gold Rush apple #10-#15 Antonovka $55 2 apple Antique apple Royal Limber + Pomme Gris #10-#15 Domestic $65 1 double graft apple apple Antique apple Winter Banana heritage apple #10-#15 Domestic $55 3 apple Page 1 of 4 -

Vyhláška Č. 331/2017 Sb
zakonyprolidi_cs_2017_331_v20180124 https://www.zakonyprolidi.cz/print/cs/2017-331/zneni-20180124.htm Vyhláška č. 331/2017 Sb. Vyhláška o stanovení dalších odrůdovocných druhů s úředně uznaným popisem, které se považují za zapsané do Státní odrůdové knihy https://www.zakonyprolidi.cz/cs/2017-331 Částka 113/2017 Platnost od 11.10.2017 Účinnost od 01.11.2017 Aktuální znění 24.01.2018 331 VYHLÁŠKA ze dne 2. října 2017 o stanovení dalších odrůd ovocných druhů s úředně uznaným popisem, které se považují za zapsané do Státní odrůdové knihy Ministerstvo zemědělství stanoví podle § 35c odst. 5 zákona č. 219/2003 Sb., o uvádění do oběhu osiva a sadby pěstovaných rostlin a o změně některých zákonů (zákon o oběhu osiva a sadby), ve znění zákona č. 295/2017 Sb.: § 1 Další odrůdy ovocných druhů s úředně uznaným popisem, které se považují za zapsané do Státní odrůdové knihy, jsou uvedeny v příloze k této vyhlášce. § 2 Účinnost Tato vyhláška nabývá účinnosti dnem 1. listopadu 2017. Ministr: Ing. Jurečka v. r. Příloha k vyhlášce č. 331/2017 Sb. Seznam dalších odrůd ovocných druhů s úředně uznaným popisem, které se považují za zapsané do Státní odrůdové knihy Druh Odrůda Líska (Corylus avellana L.) Lombardská červená Římský Kdouloň (Cydonia oblonga Milí.) Asenica Bereczkého Hruškovitá Izobilnaja Kocurova Leskovačka Muškatnaja Selena Jahodník (Fragaria L.) Evita Frikonsa Kama 1 z 11 07.03.2018, 13:22 zakonyprolidi_cs_2017_331_v20180124 https://www.zakonyprolidi.cz/print/cs/2017-331/zneni-20180124.htm Lesana Maranell Mount Everest Olivie Polka Roxana Vanda -
Edible Landscape Nursery at Central Rocky Mountain Permaculture Institute (CRMPI) Fruit/ Common Name Rootstock Price Available Function
Edible Landscape Nursery at Central Rocky Mountain Permaculture Institute (CRMPI) Fruit/ Common Name Rootstock Price Available Function Apple Akane + Greening double graft Domestic apple $65 1 apple Apple Akane apple Domestic apple $55 1 Apple Alexander apple $55 2 Apple Arkcharm apple $45 1 Apple E. German Round apple Antonovka $55 5 apple Apple Gala apple M7 semi-dwarf $55 1 apple Apple Ginger Gold apple Domestic apple $55 1 Apple Green Rabbit apple Domestic apple $55 3 Apple Greening apple Domestic apple $55 1 Apple Honeycrisp apple M7 semi-dwarf $55 1 Apple Lodi apple Domestic apple $55 4 Apple Lord's Seedling apple Domestic apple $55 1 Apple Mac Fence apple $55 1 Apple McIntosh + Greening double Domestic apple $65 1 graft apple Apple Mutsu apple M111 semi- $55 1 dwarf apple Apple Mystery apple $45 5 Apple Mystery apple double graft Domestic apple $55 1 Apple Nanette apple Antonovka $55 3 apple Apple New Mac apple Antonovka $55 3 apple 05/09/2017 Page 1 of 5 Fruit/ Common Name Rootstock Price Available Function Apple New Mac apple Domestic apple $55 2 Apple Northern Spy apple M111 semi- $55 1 dwarf apple Apple Red Baron apple Domestic apple $55 1 Apple Red Gravenstein apple M111 semi- $50 5 dwarf apple Apple Sweet Sixteen apple Antonovka $55 3 apple Apple Yellow transparent apple Antonovka $55 1 apple Apple Wolf River + Gold Rush apple Antonovka $75 1 double graft apple Crabapple Kerr flowering crabapple $45 1 Crabapple Mystery crabapple $45 1 Crabapple Whitney crabapple M7 semi-dwarf $55 1 apple Crabapple Whitney crabapple Domestic -

Audzēšanai Ieteicamās Ābeļu Škirnes
Audzēšanai ieteicamās ābeļu šķirnes Laila Ikase Latvijas Valsts augļkopības institūts www.lvai.lv Šķirnes izvēles kritēriji • 1. Dārza vieta un augsne. • 2. Šķirnes ziemcietība un slimībizturība. • 3. Piemērotība audzētāja vajadzībām: – pašu patēriņam, – tirgum (uz vietas vai tālākpārdošanai), – pārstrādei. • 4. Kopšanas un novākšanas iespējas. • 5. Glabāšanas iespējas. ? Vai zināms, kā šķirne uzvedas kaimiņu dārzos? 1. ŠĶIRNES KOMERCDĀRZIEM (Var audzēt arī mazdārzos) VASARAS ŠĶIRNES Šķirnes Raksturīgās īpašības Priekšrocības Problēmas Agra Agra vasaras, augļi Augļi – ienākas pirms ‘Baltā Augļi – neretinot var būt pasīki, vidēji lieli, sarkani, Dzidrā’, izskatīga krāsa. iespējams rūsinājums, glabājot vīst. saldskābi. Koks – ražīgs, slimībizturīgs, Koks – ražas kāpums diezgan lēns, ziemcietīgs. vainagam tieksme sabiezināties. Baltais Dzidrais Agra vasaras, augļi Augļi – izskatīgi, retinot lieli. Augļi – zema transportizturība, īss vidēji līdz lieli, bāli glabāšanas laiks, labam lielumam Koks – pieticīgs, ražīgs, dzelteni, saldskābi. jāretina. pietiekami izturīgs pret kraupi. Laba pārstrādei. Koks – ražo pārgadus, vidēja izturība pret vēzi. Konfetnoje Vasaras, augļi vidēji lieli, Augļi – ļoti garšīgi, saldi. Augļi – vācami pakāpeniski, labam viegli svītraini, saldi ar krāsojumam jāvāc tikai pilnīgi gatavi. Koks – ziemcietīgs, izturīgs pret vieglu skābumu. slimībām. Koks – ražo pārgadus, ražas kāpums palēns. Krapes Vasaras, nedaudz vēlāk Augļi – ļoti garšīgi, saldi. Augļi – vācami pakāpeniski, labam Cukuriņš par Konfetnoje, augļi krāsojumam jāvāc tikai pilnīgi gatavi. Koks – ziemcietīgs, izturīgs pret ļoti līdzīgi. slimībām. Koks – ražo pārgadus, tieksme veidot Koks spēcīgāka auguma stāvu vainagu. nekā ‘Konfetnoje’. ‘Agra’ ‘Baltais Dzidrais’ ‘Konfetnoje’ ‘Krapes Cukuriņš’ RUDENS – AGRAS ZIEMAS ŠĶIRNES Šķirnes Raksturīgās īpašības Priekšrocības Problēmas Kovaļenkov- Vēla vasaras – rudens, Augļi – izskatīgi, saldi. Augļi – glabājot zaudē kvalitāti, bez skoje augļi lieli, sarkani, saldi vainaga izgaismošanas slikts krāsojums. Koks – labi ražo jebkuros bez skābuma. -

Poster Session Abstracts POSTERS–Saturday 106Th Annual International Conference of the American Society for Horticultural Science Millennium Hotel, St
Poster Session Abstracts POSTERS–Saturday 106th Annual International Conference of the American Society for Horticultural Science Millennium Hotel, St. Louis, Missouri All posters are displayed in the Poster Hall, located in the Illinois/Missouri/Meramac rooms. The number in parentheses ( ) preceding the poster title corresponds to the location of the poster within the Poster Hall. Presenting authors are denoted by an asterisk (*). (42) Rheological Properties of Water-soluble Crop Physiology/Physiology: Polysaccharide in Peach Gum from Cross-Commodity Almond (Prunus dulcis) Saturday, 25 July 2009 12:00–12:45 pm Sen Wang Central South University of Forestry and Technology, Changsha; wangq- (41) Growth and Salinity Tolerance of Zinnia elegans [email protected] When Irrigated with Wastewater from Two Distinct Lin Zhang Central South University of Forestry and Technology, Huann 410004; Growing Regions in California [email protected] Christy T. Carter* Deyi Yuan* Tennessee Tech University, Cookeville, TN; [email protected] Central South University of Forestry and Technology, Changsha; yuan- Catherine Grieve [email protected] U.S. Salinity Laboratory, Riverside, CA; [email protected] Qiuping Zhong Using recycled greenhouse effluents to irrigate salt-tolerant floral crops Central South University of Forestry and Technology, Changsha; wangq- provides an economic and environmental benefit for growers. Produc- [email protected] ers are able to reduce their direct use of high quality water for certain Yina Li crops and simultaneously reduce or prevent groundwater contamina- Central South University of Forestry and Technology, Changsha; wangq- tion. We selected Zinnia elegans as a potential salt-tolerant crop for [email protected] use in a recycled greenhouse system based on the known hardiness of The rheological properties of water-soluble polysaccharide in peach its wild relatives and because of its economic value to the floriculture gum from almond (Prunus dulcis) were studied in this paper in order industry. -

Advances in Apple Breeding for Enhanced Fruit Quality and Resistance to Biotic Stresses: New Varieties for the European Market
PROTECTION OF GENETIC RESOURCES OF POMOLOGICAL PLANTS AND SELECTION OF GENITORS WITH TRAITS VALUABLE FOR SUSTAINABLE FRUIT PRODUCTION Journal of Fruit and Ornamental Plant Research vol. 12, 2004 Special ed. ADVANCES IN APPLE BREEDING FOR ENHANCED FRUIT QUALITY AND RESISTANCE TO BIOTIC STRESSES: NEW VARIETIES FOR THE EUROPEAN MARKET Silviero Sansavini, Franco Donati, Fabrizio Costa and Stefano Tartarini Dipartimento di Colture Arboree, Viale Fanin 46, University of Bologna, ITALY Phone: +39 0512096400 Fax: +39 0512096401 e•mail: [email protected] (Received November 5, 2004/Accepted January 20, 2005) ABSTRACT The current trends and future prospects for apple breeding in the newly enlarged 25•member European Union (EU) are reported. The last twenty years have seen a marked rise in both the number of breeding programmes and of the cultivars they have released. The main objectives of these efforts have focused on resistance to diseases like scab, mildew and fire blight and on enhanced fruit quality in its broadest sense•appearance, sensory traits, storability and shelf•life. While there are many new scab•resistant apples, their appeal to mainstream consumers is notably restricted. Yet the future appears to hold much promise for these programmes. The use of new biotech tools should accelerate the development of novel varieties while saving time and reducing work loads. Indeed, several stations have already introduced marker• assisted selection (MAS) for monogenic traits and QTLs are increasingly important in segregating polygenic traits. Expectations are high that with efforts like the EU’s new HiDRAS Project traits involved in fruit quality (ripening, softening, acids, sugars, flavour, polyphenols and other antioxidant compounds) can be controlled. -

Apple Cultivars of the All-Russian Research Institute of Horticultural Breeding
SCIENTIFIC WORKS OF THE LITHUANIAN INSTITUTE OF HORTICULTURE AND LITHUANIAN UNIVERSITY OF AGRICULTURE. SODININKYSTĖ IR DARŽININKYSTĖ. 2008. 27(4). Apple cultivars of the All-Russian Research Institute of Horticultural Breeding Eugeny Sedov, Zoja Serova, Nina Krasova The All-Russian Research Institute of Horticultural Breeding (VNIISPK) Russia, Orel, p/o Zhilina, e-mail: [email protected] The results of over semi-centennial apple breeding in the All-Russian Research Institute of Horticultural Breeding are shown. The brief descriptions of 33 apple cultivars are given, in particular 8 summer cultivars, 3 autumn ones and 22 cultivars of winter maturing. Most cultivars have been included into the State Register of Breeding Achievements admitted to cultivation in Russian Federation, the other are on State trials. Key words: trees, fruit, breeding, cultivars, resistance, apple. Introduction. The requirements to apple cultivars have increased significantly in connection with the intensification of horticulture. Many old middle Russian cul - tivars of folk selection, cultivars of I. V. Michurin and his followers I. S. Gorshkov, S. F. Chernenko, S. I. Isaev, S. P. Kedrin and others played their important part in assortment and creating new cultivars. In the central part of Russia the All-Russian Research Institute of Horticultural Breeding (VNIISPK) (former Orel zone fruit-berry experiment station) is the basic supplier of new apple cultivars corresponding to the modern requirements. The Institute is the oldest pomological institution of Russia, and in 2005 it celebrated its 160 th anniversary. The cultivars developed in this Institute are the base of the regionalized apple assortment in the Central-Chernozem area (Belgorod, Voronezh, Kursk, Lipetsk, Orel, Tambov and other regions), Central area (Bryansk, Kaluga, Moscow, Ryazan, Tula and other regions) and some other regions and areas of Russia (Sedov, Krasova, 2000; Sedov, 1999; Sedov et al., 2004; Sedov, 2005, 2006 a, 2006 b, 2007 a, 2007 b, 2007 c). -

Tree Physical Characteristics 1 Variety Vigor 36-305 Adam's Pearmain
Tree Physical Characteristics 1-few cankers 2- moderate (less than 10 lesions), non lethal 3- girdling, lethal or g-50-90% 1 - less than 5% extensive a- f-35-50% 2 - 5-15% anthracnose canker p -less than 35% 3 - More than 15% 3-strongest 1 yes or no n- nectria (from vertical) 4 - severe (50% ) alive or dead 1-weakest branch dieback/cold 1 Variety leaf scab canker angle damage mortality notes vigor 36-305 y a 1 g 1 a perfect form 3 1 weak, 1 decent, spindly Adam's Pearmain n a 1 p 2 a growth 2 Adanac n 0 f 3 a weak 1 no crop in 11 Akero y f a years 2 Albion n 0 f 0 a 2 only 10% of fruit had scab, none on leaves, Alexander n 0 f a bullseye on fruit 3 bad canker, Alkmene y a 1,3 f 0 a bushy 2 vigorous but Almata y f 0 a lanky 3 red 3/4 fruits, nice open form, Almey y 0 g 1 a severe scab 3 Altaiski Sweet y a Alton y g 1 a 3 American Beauty y f 0 a slow to leaf out 3 Amsib Crab n f 1 a 2 looks decent, Amur Deburthecort y 2 a cold damage early leaf drop, Anaros y 2 g 0 a nicely branched 3 dieback spurs Anis n a 1 , other cankers p 1 a only 1 Anise Reinette y a 1 f 2 a 2 Anisim y a 1 p 1 a leader dieback 3 Ann's n 0 f 2 a local variety 2 Anoka y f a 2 Antonovka 81 n g a 3 Antonovka 102 n g a 3 Antonovka 109 n f a 3 antonovka 52 n g a 3 Antonovka 114 n g a 3 Antonovka 1.5 n a1 g 0 a 3 n f 0 a 3 Antonovka 172670-B Page 1 of 17 Tree Physical Characteristics Antonovka 37 n g a 3 Antonovka 48 n g a 3 Antonovka 49 n g a 3 Antonovka 54 n f a slow to leaf out 3 Antonovka Debnicka n f 0 a 3 Antonovka Kamenichka n a 1 f 0 a 3 Antonovka Michurin n 0 f -

Dihydrochalcones in Malus Mill. Germplasm and Hybrid
DIHYDROCHALCONES IN MALUS MILL. GERMPLASM AND HYBRID POPULATIONS A Dissertation Presented to the Faculty of the Graduate School of Cornell University In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by Benjamin Leo Gutierrez December 2017 © 2017 Benjamin Leo Gutierrez DIHYDROCHALCONES IN MALUS MILL. GERMPLASM AND HYBRID POPULATIONS Benjamin Leo Gutierrez, Ph.D. Cornell University 2017 Dihydrochalcones are abundant in Malus Mill. species, including the cultivated apple (M. ×domestica Borkh.). Phloridzin, the primary dihydrochalcone in Malus species, has beneficial nutritional qualities, including antioxidant, anti-cancer, and anti-diabetic properties. As such, phloridzin could be a target for improvement of nutritional quality in new apple cultivars. In addition to phloridzin, a few rare Malus species produce trilobatin or sieboldin in place of phloridzin and hybridization can lead to combinations of phloridzin, trilobatin, or sieboldin in interspecific apple progenies. Trilobatin and sieboldin also have unique chemical properties that make them desirable targets for apple breeding, including high antioxidant activity, anti- inflammatory, anti-diabetic properties, and a high sweetness intensity. We studied the variation of phloridzin, sieboldin, and trilobatin content in leaves of 377 accessions from the USDA National Plant Germplasm System (NPGS) Malus collection in Geneva, NY over three seasons and identified valuable genetic resources for breeding and researching dihydrochalcones. From these resources, five apple hybrid populations were developed to determine the genetic basis of dihydrochalcone variation. Phloridzin, sieboldin, and trilobatin appear to follow segregation patterns for three independent genes and significant trait-marker associations were identified using genetic data from genotyping-by-sequencing. Dihydrochalcones are at much lower quantities in mature apple fruit compared with vegetative tissues. -

LOW TEMPERATURE TOLERANCE of APPLE CULTIVARS of DIFFERENT PLOIDY at DIFFERENT TIMES of the WINTER Zoya Ozherelieva# and Evgeny Sedov
PROCEEDINGS OF THE LATVIAN ACADEMY OF SCIENCES. Section B, Vol. 71 (2017), No. 3 (708), pp. 127–131. DOI: 10.1515/prolas-2017-0022 LOW TEMPERATURE TOLERANCE OF APPLE CULTIVARS OF DIFFERENT PLOIDY AT DIFFERENT TIMES OF THE WINTER Zoya Ozherelieva# and Evgeny Sedov Federal State Budget Scientific Institution All Russian Research Institute of Fruit Crop Breeding (FGBNU VNIISPK), 302530 Orel region, Zhilina, RUSSIA # Corresponding author, [email protected] Communicated by Edîte Kaufmane Artificial freezing was used to evaluate diploid and triploid apple cultivars from the All Russian Re- search Institute of Fruit Crop Breeding at Orel throughout three winters. The studied apple variet- ies were developed by breeder E. N. Sedov and cytological analysis was carried out by cytologist G. A. Sedysheva. In early winter, all cultivars exhibited high tolerance to cold. In mid-winter buds and wood were severely damaged, while bark was more resistant for most cultivars. Basic com- ponents of hardiness were estimated: component I — frost resistance at –25 °C in the beginning of winter; component II — maximum value of frost resistance at –40 °C developed by plants dur- ing hardening; component III — ability to retain the hardened condition at –25 °C after a period of three-day thaw at +2 °C; and component IV — the ability to restore frost resistance at –30 °C af- ter repeated hardening and three-day thaw at +2 °C. During late-winter thaws, buds suffered from frosts, while the bark and wood retained frost hardiness. Late in winter all cultivars demonstrated high resistance to repeated frosts. Triploid cultivars exhibited the highest level of cold hardiness of vegetative buds, bark and wood of annual shoots throughout the winter; these cultivars in- cluded ‘Zhilinskoye’, ‘Vavilovskoye’, ‘Osipovskoye’, ‘Patriot’, ‘Sinap Orlovski’, ‘Spasskoye’, ‘Turgenevskoye’, and diploids ‘Bolotovskoye’, ‘Sokovinka’, and ‘’Ranneye Aloye’. -

Portland Nursery Buyer's Fruit Tree List for 2021
Portland Nursery Buyer’s Fruit Tree List for 2021 Apple root stocks Cherry rootstock Figs Akane m26, m27 Bing gisela,newroot,mah Black Mission Arkansas Black m106 Black Gold gisela Black Spanish Ashmead's Kernel m26 Black Tartarian mahaleb Brown Turkey Belle de Boskoop m111 Carmine Jewel gisela, own-root Chicago Hardy Bramley m106 Combination, 4-way gisela, mazzard Excell Caville Blanc m111 Corum mahaleb King Chehalis m106 Evans gisela Lattarula Combination, 4-way m106 Glacier gisela Negronne Cox Orange Pippin m26,m106 Lapin gisela,mahaleb Olympian Early Pink Lady m26 Meteor mahaleb Oregon Prolific Empire m106 Montmorency gisela,mahaleb Panache Enterprise m106 North Star mahaleb Peter's Honey Espalier, 2-tier or 3-tier m106 Rainier gisela,newroot,mah Petite Negra Freedom m26 Royal Ann gisela Stella Fuji m27,m26,m106 Sam gisela Vern's Brown Turkey Gala m27,m26 Stella gisela,mahaleb Granny Smith gen935,m106 Sweetheart gisela,newroot,mah Olive Gravenstein m27,m26 Van gisela Arbequina Haralson antonovka Vandelay gisela Honeycrisp m27,m26,gen935 Pawpaw King David m111 Plum Mango Liberty m27,m26 Beauty citation,mar2624 Overleese Melrose m26,m106 Blue Damson st. julian a Shenandoah Northern Spy m106 Brooks mar2624 Pink Lady m27,m26 Burgundy citation Mulberry Pink Pearl m26 Catalina citation Beautiful Day Pristine m26 Combination, 4-way krymsk,mar2624 Illinois Everbearing Red McIntosh m26 Early Italian mar2624 Oscar Roxbury Russet m106 Elephant Heart mar2624 Pakistan Spartan m106 French Petite mar2624 Teas Weeping Spitzenberg m106 Green Gage mar2624 William's Pride m26 Hollywood citation Persimmon Winesap m106 Imperial Epineuse st. julian a Fuyu Yellow Newton m106 Italian citation, mar2624 Yellow Transparent m106 Methley mar2624 Nuts Nadia st. -

PENNSYLVANIA 2012–2013 Tree Fruit Production Guide
PENNSYLVANIA 2012–2013 Tree Fruit Production Guide College of Agri C u lt u r A l S C i e n C e S Production Guide Pesticide Safety Coordinator Penn State Pesticide Education J. M. Halbrendt Program Horticulture Orchard Sprayer Techniques R. M. Crassweller, J. W. Travis section coordinator Harvest and Postharvest Handling J. R. Schupp R. M. Crassweller Entomology J. R. Schupp G. Krawczyk, section Cider Production and Food Safety coordinator L. F. LaBorde L. A. Hull D. J. Biddinger Orchard Budgets J. K. Harper Pollination and Bee L. F. Kime Management M. Frazier Farm Labor Regulations D. J. Biddinger R. H. Pifer Plant Pathology Designer H. K. Ngugi, section G. Collins coordinator Editor N. O. Halbrendt A. Kirsten Nematology Disease, insect figures J. M. Halbrendt C. Gregory Wildlife Resources C. Jung G. San Julian Cover Photos Environmental Monitoring iStock J. W. Travis this guide is also available on the web at agsci.psu.edu/tfpg Visit Penn State’s College of Agricultural Sciences on the web: agsci.psu.edu This publication is available from the Publications Distribution Center, The Pennsylvania State Univer- sity, 112 Agricultural Administration Building, University Park, PA 16802. For information telephone 814-865-6713. Where trade names appear, no discrimination is intended, and no endorsement by Penn State Cooperative Extension or the College of Agricultural Sciences is implied. This publication is available in alternative media on request. The Pennsylvania State University is committed to the policy that all persons shall have equal access to programs, facilities, admission, and employment without regard to personal characteristics not related to ability, performance, or qualifications as determined by University policy or by state or federal authorities.