(Rosaceae) Based on Integrated Evidence from Nuclear and Plastid Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Malosma Laurina (Nutt.) Nutt. Ex Abrams
I. SPECIES Malosma laurina (Nutt.) Nutt. ex Abrams NRCS CODE: Family: Anacardiaceae MALA6 Subfamily: Anacardiodeae Order: Sapindales Subclass: Rosidae Class: Magnoliopsida Immature fruits are green to red in mid-summer. Plants tend to flower in May to June. A. Subspecific taxa none B. Synonyms Rhus laurina Nutt. (USDA PLANTS 2017) C. Common name laurel sumac (McMinn 1939, Calflora 2016) There is only one species of Malosma. Phylogenetic analyses based on molecular data and a combination of D. Taxonomic relationships molecular and structural data place Malosma as distinct but related to both Toxicodendron and Rhus (Miller et al. 2001, Yi et al. 2004, Andrés-Hernández et al. 2014). E. Related taxa in region Rhus ovata and Rhus integrifolia may be the closest relatives and laurel sumac co-occurs with both species. Very early, Malosma was separated out of the genus Rhus in part because it has smaller fruits and lacks the following traits possessed by all species of Rhus : red-glandular hairs on the fruits and axis of the inflorescence, hairs on sepal margins, and glands on the leaf blades (Barkley 1937, Andrés-Hernández et al. 2014). F. Taxonomic issues none G. Other The name Malosma refers to the strong odor of the plant (Miller & Wilken 2017). The odor of the crushed leaves has been described as apple-like, but some think the smell is more like bitter almonds (Allen & Roberts 2013). The leaves are similar to those of the laurel tree and many others in family Lauraceae, hence the specific epithet "laurina." Montgomery & Cheo (1971) found time to ignition for dried leaf blades of laurel sumac to be intermediate and similar to scrub oak, Prunus ilicifolia, and Rhamnus crocea; faster than Heteromeles arbutifolia, Arctostaphylos densiflora, and Rhus ovata; and slower than Salvia mellifera. -

Plum Crazy: Rediscovering Our Lost Prunus Resources W.R
Plum Crazy: Rediscovering Our Lost Prunus Resources W.R. Okie1 U.S. Department of Agriculture–Agricultural Research Service, Southeastern Fruit and Tree Nut Research Laboratory, 21 Dunbar Road, Byron, GA 31008 Recent utilization of genetic resources of peach [Prunus persica (‘Quetta’ from India, ‘John Rivers’ from England, and ‘Lippiatts’ (L.) Batsch] and Japanese plum (P. salicina Lindl. and hybrids) has from New Zealand) were critical to the development of modern been limited in the United States compared with that of many crops. nectarines in California (Taylor, 1959). However, most fresh-market Difficulties in collection, importation, and quarantine throughput have peach breeding programs in the United States have used germplasm limited the germplasm available. Prunus is more difficult to preserve developed in the United States for cultivar development (Okie, 1998). because more space is needed than for small fruit crops, and the shorter Only in New Jersey was there extensive hybridization with imported life of trees relative to other tree crops because of disease and insect clones, and most of these hybrids have not resulted in named cultivars problems. Lack of suitable rootstocks has also reduced tree life. The (Blake and Edgerton, 1946). trend toward fewer breeding programs, most of which emphasize In recent years, interest in collecting and utilizing novel germplasm “short-term” (long-term compared to most crops) commercial cultivar has increased. For example, non-melting clingstone peaches from development to meet immediate industry needs, has also contributed Mexico and Brazil have been used in the joint USDA–Univ. of to reduced use of exotic material. Georgia–Univ. of Florida breeding program for the development of Probably all modern commercial peaches grown in the United early ripening, non-melting, fresh-market peaches for low-chill areas States are related to ‘Chinese Cling’, a peach imported from China (Beckman and Sherman, 1996). -

Prunus Ilicifolia (Hooker & Arnott) D. Dietr. Subsp. Ilicifolia, HOLLY
Prunus ilicifolia (Hooker & Arnott) D. Dietr. subsp. ilicifolia , HOLLY-LEAVED CHERRY, ISLAY . Shrub or small tree, evergreen, sclerophyllous, somewhat spinescent, highly branched and forming a dense canopy, 100–800 cm tall; shoots with dark green foliage, ± folded upward from midrib, with leaves bent downward and hiding stem on water-stressed shrubs, essentially glabrous. Stems: cylindric, when young reddish brown to grayish brown, with odor of bitter almonds (hydrogen cyanide) when scratched; old bark grayish, fairly smooth, with small horizontal lenticels. Leaves: helically alternate, simple, petiolate, with stipules; stipules 2, attached to petiole base, awl-shaped, < 2.5 mm long, entire or sometimes fringed, with some hairs near base, aging purple and early-deciduous; petiole 4–11 mm long, glands absent; blade ovate to round, 16–65 × 12–50 mm, tough, rounded to cordate at base, spinose-dentate or serrate on margins and often wavy, obtuse to rounded or truncate at tip, pinnately veined with midrib raised on lower surface, becoming glabrescent, upper surface initially glossy aging satiny. Inflorescence: raceme or panicle with a central raceme and 2–4 lateral racemes from base, arising mostly from dormant bud prior to vegetative growth, having bud scales at base, each raceme 13–30- flowered, (15–)25–80 × 13–20 mm, flowers ± alternate, bracteate, glabrous to glabrate; bract subtending raceme leaflike but reduced or broadly triangular and 3-toothed at tip; rachis somewhat ridged, glabrous or with widely scattered hairs; bractlet subtending pedicel ± triangular to awl-shaped or cupped-ovate, 0.7–2.6 mm long, light green, deeply 3-toothed with acuminate teeth (basal flowers) to acuminate at tip, sparsely short-ciliate mostly above midpoint, abscising when bud small leaving a raised scar; pedicel 0.2–5 mm long. -

Habitat Guidelines for Mule Deer: California Woodland Chaparral Ecoregion
THE AUTHORS : MARY L. SOMMER CALIFORNIA DEPARTMENT OF FISH AND GAME WILDLIFE BRANCH 1812 NINTH STREET SACRAMENTO, CA 95814 REBECCA L. BARBOZA CALIFORNIA DEPARTMENT OF FISH AND GAME SOUTH COAST REGION 4665 LAMPSON AVENUE, SUITE C LOS ALAMITOS, CA 90720 RANDY A. BOTTA CALIFORNIA DEPARTMENT OF FISH AND GAME SOUTH COAST REGION 4949 VIEWRIDGE AVENUE SAN DIEGO, CA 92123 ERIC B. KLEINFELTER CALIFORNIA DEPARTMENT OF FISH AND GAME CENTRAL REGION 1234 EAST SHAW AVENUE FRESNO, CA 93710 MARTHA E. SCHAUSS CALIFORNIA DEPARTMENT OF FISH AND GAME CENTRAL REGION 1234 EAST SHAW AVENUE FRESNO, CA 93710 J. ROCKY THOMPSON CALIFORNIA DEPARTMENT OF FISH AND GAME CENTRAL REGION P.O. BOX 2330 LAKE ISABELLA, CA 93240 Cover photo by: California Department of Fish and Game (CDFG) Suggested Citation: Sommer, M. L., R. L. Barboza, R. A. Botta, E. B. Kleinfelter, M. E. Schauss and J. R. Thompson. 2007. Habitat Guidelines for Mule Deer: California Woodland Chaparral Ecoregion. Mule Deer Working Group, Western Association of Fish and Wildlife Agencies. TABLE OF CONTENTS INTRODUCTION 2 THE CALIFORNIA WOODLAND CHAPARRAL ECOREGION 4 Description 4 Ecoregion-specific Deer Ecology 4 MAJOR IMPACTS TO MULE DEER HABITAT 6 IN THE CALIFORNIA WOODLAND CHAPARRA L CONTRIBUTING FACTORS AND SPECIFIC 7 HABITAT GUIDELINES Long-term Fire Suppression 7 Human Encroachment 13 Wild and Domestic Herbivores 18 Water Availability and Hydrological Changes 26 Non-native Invasive Species 30 SUMMARY 37 LITERATURE CITED 38 APPENDICIES 46 TABLE OF CONTENTS 1 INTRODUCTION ule and black-tailed deer (collectively called Forest is severe winterkill. Winterkill is not a mule deer, Odocoileus hemionus ) are icons of problem in the Southwest Deserts, but heavy grazing the American West. -

The Pubescent-Fruited Species of Prunus of the Southwestern States
THE PUBESCENT-FRUITED SPECIES OF PRUNUS OF THE SOUTHWESTERN STATES By SIMS C. MASON, Arboriculturist, Crop Physiology and Breeding Investigations, Bureau of Plant Industry INTRODUCTION The species of the genus Prunus described in this article occupy a unique position in the flora of the western United States from the fact that their relationship with the wild plums of the country is remote and they are more closely allied to some of the Asiatic species of this genus. Their economic importance arises chiefly from their close adaptation to the climatic and soil conditions of the Southwest, where fluctuations of heat and cold, severe drought, and considerable alkalinity of the soil must be endured by most tree crops. Adaptable stocks for the cultivated forms of Prunus capable of meeting such conditions are eagerly sought. Species with such characters which are capable of being hybridized with the old-established cultivated forms of the genus offer attractive possibilities to the plant breeder. This is especially true of the one edible-fruited form, Prunus texana, which affords in aroma and flavor of fruit most attractive characters for combi- nation with other stone fruits of larger size and more staple commercial character. Instead of forming a homogeneous group, as has usually been be- lieved, these species fall into small groups of quite diverse character and affinities. To the plant breeder and student of their economic possibilities these relationships are of such importance that the following detailed study of them is deemed essential -

Taming the Wild Beach Plum
Taming the Wild Beach Plum Richard H. Uva Beach plum is a conspicuous shrub of coastal plant communities in the north- eastern United States because of its prolific bloom, prized fruit, and perseverance in a seemingly hostile environment. Several attempts have been made to bring this wild fruit into cultivation. ’ve known the beach plum (Prunus maritima Marsh.) I since childhood on Cape Cod, where it was the only woody plant in the sea of dune grass that sepa- rated the ocean from the rest of the world. Michael Dirr writes in his Manual of Cultivated Plants that “This species abounds on Cape Cod, Massachusetts, and is one of the Cape Codder’s cher- ALL PHOTOGRAPHS ARE BY THE AUTHOR ished plants.” In fact, I would say that Cape Codders feel a sense of entitlement to the species and its fruit. The beach plum is much appreciated for its profuse white bloom in spring, but it is in late summer, when people gather the fruit from the wild for jelly and other preserves, that its impor- The fruits of Prunus maritima are small plums—one-half to an inch (1.5 to tance to the local culture becomes 2.5 cm) in diameter—that ripen from late August through September. most apparent. The long-time gatherers have secret spots and favorite bushes, breviligulata), beach pea (Lathyrus maritimus), and strangers carrying pails in the dunes are and seaside goldenrod (Solidago sempervirens). viewed with suspicion. In a good crop year the When I learned that William Clark of the Cape race to harvest is so competitive that the fruit Cod Cooperative Extension and a small group of is sometimes picked when barely ripe. -

Overwintering Hosts for the Exotic Leafroller
Overwintering Hosts for the Exotic Leafroller Parasitoid, Colpoclypeus florus: Implications for Habitat Manipulation to Augment Biological Control of Leafrollers in Pome Fruits Author(s): R. S. Pfannenstiel, T. R. Unruh and J. F. Brunner Source: Journal of Insect Science, 10(75):1-13. 2010. Published By: Entomological Society of America DOI: http://dx.doi.org/10.1673/031.010.7501 URL: http://www.bioone.org/doi/full/10.1673/031.010.7501 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Journal of Insect Science: Vol. 10 | Article 75 Pfannenstiel et al. Overwintering hosts for the exotic leafroller parasitoid, Colpoclypeus florus: Implications for habitat manipulation to augment biological control of leafrollers in pome fruits R. S. Pfannenstiel1,2,3a, T. R. Unruh2, and J. F. Brunner1 1Tree Fruit Research and Extension Center, Washington State University, 1100 N. -

Native Plant List CITY of OREGON CITY 320 Warner Milne Road , P.O
Native Plant List CITY OF OREGON CITY 320 Warner Milne Road , P.O. Box 3040, Oregon City, OR 97045 Phone: (503) 657-0891, Fax: (503) 657-7892 Scientific Name Common Name Habitat Type Wetland Riparian Forest Oak F. Slope Thicket Grass Rocky Wood TREES AND ARBORESCENT SHRUBS Abies grandis Grand Fir X X X X Acer circinatumAS Vine Maple X X X Acer macrophyllum Big-Leaf Maple X X Alnus rubra Red Alder X X X Alnus sinuata Sitka Alder X Arbutus menziesii Madrone X Cornus nuttallii Western Flowering XX Dogwood Cornus sericia ssp. sericea Crataegus douglasii var. Black Hawthorn (wetland XX douglasii form) Crataegus suksdorfii Black Hawthorn (upland XXX XX form) Fraxinus latifolia Oregon Ash X X Holodiscus discolor Oceanspray Malus fuscaAS Western Crabapple X X X Pinus ponderosa Ponderosa Pine X X Populus balsamifera ssp. Black Cottonwood X X Trichocarpa Populus tremuloides Quaking Aspen X X Prunus emarginata Bitter Cherry X X X Prunus virginianaAS Common Chokecherry X X X Pseudotsuga menziesii Douglas Fir X X Pyrus (see Malus) Quercus garryana Garry Oak X X X Quercus garryana Oregon White Oak Rhamnus purshiana Cascara X X X Salix fluviatilisAS Columbia River Willow X X Salix geyeriana Geyer Willow X Salix hookerianaAS Piper's Willow X X Salix lucida ssp. lasiandra Pacific Willow X X Salix rigida var. macrogemma Rigid Willow X X Salix scouleriana Scouler Willow X X X Salix sessilifoliaAS Soft-Leafed Willow X X Salix sitchensisAS Sitka Willow X X Salix spp.* Willows Sambucus spp.* Elderberries Spiraea douglasii Douglas's Spiraea Taxus brevifolia Pacific Yew X X X Thuja plicata Western Red Cedar X X X X Tsuga heterophylla Western Hemlock X X X Scientific Name Common Name Habitat Type Wetland Riparian Forest Oak F. -

THE POL&IHATXON STATUS of PRUNUS SUBCQRDATA by LEWIS
THE POL&IHATXON STATUS OF PRUNUS SUBCQRDATA by LEWIS ANGLE BAMMEfiS A THESIS submittod to 0KEO0M STATE COLt«EGB in partial fulfillsont of the requirements for the degree of MASTER OF SCIBSCI June 1949 APPROVED: Head of pe^artment of Horticulture In Charge of Major ... i tyf fin' i in — w—r i ,.,... i Chairman of School Graduate Committee ' -*• .~.- -— ^. ^. ^^_^y^ Dean of Graduate School 7— 3" ; ACKNOWLEDGEMENTS The author wishes to express his grateful appre- ciation for the valuable assistance rendered by the many persons cooperating in this atudy* He is particularly Indebted to Professor Henry Hartmen, whose guidance and generous help carried this work to its completion. To A* H. Roberts, Assistant Horticulturaliat of the Experi- ment Station, the author is deeply indebted for helpful assistance in all phases of the problem without which the study would not have reached a successful conclusion, L. A, H. Tmm OF CONTESTS Paga Introduction ..*.......... 1 History ............ 1 Burbank^s work with the wild plum » ... 1 Sisson plvtm ............ 2 Botanical Bascription * . ........ 4 Original desorlption ....»...* 4 Sargent's description. ....*.•. 4 Variety g^lloggii ....... ♦ . 9 Variety ftyegana ...... * -« . 10 Delimitations ............. 11 Variety selection and testing * . * . II Follina^ion studies ......... 11 Rootstook studies ••...••... 11 Botanical relationship and aeearate descriptions. • • . • « • • . 11 Origin of hybrid types . « . ♦ « » . * IS Seed or Timeliness of The Study •s; Development of plum in southern Oregon ... IS Orchard culture in XaUfce County . , . * 13 Distribution • . • . • . Sarly writers* reports ........ 15 Soil preference ...» « . • . 16 Most productive plants . .♦».♦. 16 Factor of location ......... 16 fruit Setting .......... 19 Processes taking place within the flower . * 19 Formation of pollen and embryo sac * .. -

Rhamnus Crocea Nutt
I.SPECIES Rhamnus crocea Nutt. and Rhamnus ilicifolia Kellogg NRCS CODE: Family: Rhamnaceae 1. RHCR Order: Rhamnales 2. RHIL Subclass: Rosidae Class: Magnoliopsida Rhamnus crocea, San Bernardino Co., 5 June 2015, A. Montalvo Rhamnus ilicifolia, Riverside Co., Rhamnus ilicifolia, Riverside Co. Note the finely serrulate leaf 10 June 2015. A. Montalvo margins. 7 March 2008. A. Montalvo A. Subspecific taxa 1. None recognized by Sawyer (2012b) or in 2019 Jepson e-Flora whereas R. pilosa is recognized as R. c. 1. RHCR Nutt. subsp. pilosa (Trel.) C.B. Wolf in the PLANTS database (USDA PLANTS 2018). 2. RHIL 2. None B. Synonyms 1. RHCR 1 . Rhamnus croceus (spelling variant noted by FNA 2018) 2. RHIL 2. Rhamnus crocea Nutt. subsp. ilicifolia (Kellogg) C.B. Wolf; R. c. Nutt. var. ilicifolia (Kellogg) Greene (USDA PLANTS 2018) last modified: 3/4/2020 RHCR RHIL, page 1 printed: 3/10/2020 C. Common name The name red-berry, redberry, redberry buckthorn, California red-berry, evergreen buckthorn, spiny buckthorn, and hollyleaf buckthorn have been used for multiple taxa of Rhamnus (Painter 2016 a,b) 1. RHCR 1. Spiny redberry (Sawyer 2012a); also little-leaved redberry (Painter 2016a) 2. RHIL 2. Hollyleaf redberry (Sawyer 2012b); also holly-leaf buckthorn, holly-leaf coffeeberry (Painter 2016b) D. Taxonomic relationships There are about 150 species of Rhamnus worldwide and 14 in North America, 6 of which were introduced from other continents (Nesom & Sawyer 2018, FNA). This is after splitting the genus into Rhamnus (buckthorns and redberries) and Frangula (coffeeberries) based on a combination of fruit, leaf venation, and flower traits (Johnston 1975, FNA). -

Little Cherry Disease: the “Little” Disease with Big Consequences
Little Cherry Disease: the “little” disease with big consequences Ken Eastwell, Plant Pathologist Clean Plant Center Northwest Washington State University - Prosser United States (2012) produced 384,646 metric tonnes with $843M farm gate value Washington produces 62% (13,759 hectares)* California produces 23% (12, 545 hectares) Oregon produces 13% (5,058 hectares) Small acreage in Michigan, Montana, Idaho, Utah, and New York *Washington yield per acre is greater than other states: Washington 7.8 tons/acre (17.4 metric tonnes/hectare) California 3.0 tons/acre (6.7 metric tonnes/hectare) Oregon 4.5 tons/acre (10.0 metric tonnes/hectare) Canada (2005) produced 6,883 metric tonnes British Columbia produces 76% (971 hectares) Ontario produces about 24% (304 hectares) Small acreage in Nova Scotia, Manitoba and Quebec Yakima County Grant County Chelan County A different pathogen involved in each case! Little cherry virus 1 (LChV1) Little cherry virus 2 (LChV2) Western X disease phytoplasma (WX) Little cherry virus 1: Family Closteroviridae Genus unassigned Includes: Grapevine leafroll associated virus 7 Mint vein banding-associated virus Little cherry virus 2 : Family Closteroviridae Thread-like particles Genus Ampelovirus Includes: Grapevine leafroll associated virus 3 Pineapple mealybug wilt-associated viruses 1 and 2 Cytopathology: Little cherry disease degrades phloem tissue and companion cells (Raine et al, Phytopathology 1976) LChV2 infected fruit is small, late ripening, poor color and insipid (low sugars and low acid) -
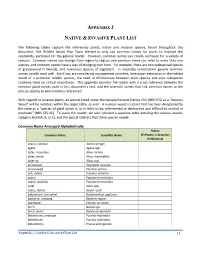
Reference Plant List
APPENDIX J NATIVE & INVASIVE PLANT LIST The following tables capture the referenced plants, native and invasive species, found throughout this document. The Wildlife Action Plan Team elected to only use common names for plants to improve the readability, particular for the general reader. However, common names can create confusion for a variety of reasons. Common names can change from region-to-region; one common name can refer to more than one species; and common names have a way of changing over time. For example, there are two widespread species of greasewood in Nevada, and numerous species of sagebrush. In everyday conversation generic common names usually work well. But if you are considering management activities, landscape restoration or the habitat needs of a particular wildlife species, the need to differentiate between plant species and even subspecies suddenly takes on critical importance. This appendix provides the reader with a cross reference between the common plant names used in this document’s text, and the scientific names that link common names to the precise species to which writers referenced. With regards to invasive plants, all species listed under the Nevada Revised Statute 555 (NRS 555) as a “Noxious Weed” will be notated, within the larger table, as such. A noxious weed is a plant that has been designated by the state as a “species of plant which is, or is likely to be, detrimental or destructive and difficult to control or eradicate” (NRS 555.05). To assist the reader, we also included a separate table detailing the noxious weeds, category level (A, B, or C), and the typical habitats that these species invade.