Multiscale Materials Modelling Programs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MODELLER 10.1 Manual
MODELLER A Program for Protein Structure Modeling Release 10.1, r12156 Andrej Saliˇ with help from Ben Webb, M.S. Madhusudhan, Min-Yi Shen, Guangqiang Dong, Marc A. Martı-Renom, Narayanan Eswar, Frank Alber, Maya Topf, Baldomero Oliva, Andr´as Fiser, Roberto S´anchez, Bozidar Yerkovich, Azat Badretdinov, Francisco Melo, John P. Overington, and Eric Feyfant email: modeller-care AT salilab.org URL https://salilab.org/modeller/ 2021/03/12 ii Contents Copyright notice xxi Acknowledgments xxv 1 Introduction 1 1.1 What is Modeller?............................................. 1 1.2 Modeller bibliography....................................... .... 2 1.3 Obtainingandinstallingtheprogram. .................... 3 1.4 Bugreports...................................... ............ 4 1.5 Method for comparative protein structure modeling by Modeller ................... 5 1.6 Using Modeller forcomparativemodeling. ... 8 1.6.1 Preparinginputfiles . ............. 8 1.6.2 Running Modeller ......................................... 9 2 Automated comparative modeling with AutoModel 11 2.1 Simpleusage ..................................... ............ 11 2.2 Moreadvancedusage............................... .............. 12 2.2.1 Including water molecules, HETATM residues, and hydrogenatoms .............. 12 2.2.2 Changing the default optimization and refinement protocol ................... 14 2.2.3 Getting a very fast and approximate model . ................. 14 2.2.4 Building a model from multiple templates . .................. 15 2.2.5 Buildinganallhydrogenmodel -

Free and Open Source Software for Computational Chemistry Education
Free and Open Source Software for Computational Chemistry Education Susi Lehtola∗,y and Antti J. Karttunenz yMolecular Sciences Software Institute, Blacksburg, Virginia 24061, United States zDepartment of Chemistry and Materials Science, Aalto University, Espoo, Finland E-mail: [email protected].fi Abstract Long in the making, computational chemistry for the masses [J. Chem. Educ. 1996, 73, 104] is finally here. We point out the existence of a variety of free and open source software (FOSS) packages for computational chemistry that offer a wide range of functionality all the way from approximate semiempirical calculations with tight- binding density functional theory to sophisticated ab initio wave function methods such as coupled-cluster theory, both for molecular and for solid-state systems. By their very definition, FOSS packages allow usage for whatever purpose by anyone, meaning they can also be used in industrial applications without limitation. Also, FOSS software has no limitations to redistribution in source or binary form, allowing their easy distribution and installation by third parties. Many FOSS scientific software packages are available as part of popular Linux distributions, and other package managers such as pip and conda. Combined with the remarkable increase in the power of personal devices—which rival that of the fastest supercomputers in the world of the 1990s—a decentralized model for teaching computational chemistry is now possible, enabling students to perform reasonable modeling on their own computing devices, in the bring your own device 1 (BYOD) scheme. In addition to the programs’ use for various applications, open access to the programs’ source code also enables comprehensive teaching strategies, as actual algorithms’ implementations can be used in teaching. -

Introducing ONETEP: Linear-Scaling Density Functional Simulations on Parallel Computers Chris-Kriton Skylaris,A) Peter D
THE JOURNAL OF CHEMICAL PHYSICS 122, 084119 ͑2005͒ Introducing ONETEP: Linear-scaling density functional simulations on parallel computers Chris-Kriton Skylaris,a) Peter D. Haynes, Arash A. Mostofi, and Mike C. Payne Theory of Condensed Matter, Cavendish Laboratory, Madingley Road, Cambridge CB3 0HE, United Kingdom ͑Received 29 September 2004; accepted 4 November 2004; published online 23 February 2005͒ We present ONETEP ͑order-N electronic total energy package͒, a density functional program for parallel computers whose computational cost scales linearly with the number of atoms and the number of processors. ONETEP is based on our reformulation of the plane wave pseudopotential method which exploits the electronic localization that is inherent in systems with a nonvanishing band gap. We summarize the theoretical developments that enable the direct optimization of strictly localized quantities expressed in terms of a delocalized plane wave basis. These same localized quantities lead us to a physical way of dividing the computational effort among many processors to allow calculations to be performed efficiently on parallel supercomputers. We show with examples that ONETEP achieves excellent speedups with increasing numbers of processors and confirm that the time taken by ONETEP as a function of increasing number of atoms for a given number of processors is indeed linear. What distinguishes our approach is that the localization is achieved in a controlled and mathematically consistent manner so that ONETEP obtains the same accuracy as conventional cubic-scaling plane wave approaches and offers fast and stable convergence. We expect that calculations with ONETEP have the potential to provide quantitative theoretical predictions for problems involving thousands of atoms such as those often encountered in nanoscience and biophysics. -

Secondary Structure Propensities in Peptide Folding Simulations: a Systematic Comparison of Molecular Mechanics Interaction Schemes
Biophysical Journal Volume 97 July 2009 599–608 599 Secondary Structure Propensities in Peptide Folding Simulations: A Systematic Comparison of Molecular Mechanics Interaction Schemes Dirk Matthes and Bert L. de Groot* Department of Theoretical and Computational Biophysics, Computational Biomolecular Dynamics Group, Max-Planck-Institute for Biophysical Chemistry, Go¨ttingen, Germany ABSTRACT We present a systematic study directed toward the secondary structure propensity and sampling behavior in peptide folding simulations with eight different molecular dynamics force-field variants in explicit solvent. We report on the combi- national result of force field, water model, and electrostatic interaction schemes and compare to available experimental charac- terization of five studied model peptides in terms of reproduced structure and dynamics. The total simulation time exceeded 18 ms and included simulations that started from both folded and extended conformations. Despite remaining sampling issues, a number of distinct trends in the folding behavior of the peptides emerged. Pronounced differences in the propensity of finding prominent secondary structure motifs in the different applied force fields suggest that problems point in particular to the balance of the relative stabilities of helical and extended conformations. INTRODUCTION Molecular dynamics (MD) simulations are routinely utilized to that study, simulations of relatively short lengths were per- study the folding dynamics of peptides and small proteins as formed and the natively folded state was used as starting well as biomolecular aggregation. The critical constituents of point, possibly biasing the results (13). such molecular mechanics studies are the validity of the under- For folding simulations, such a systematic test has not lyingphysical models together with theassumptionsofclassical been carried out so far, although with growing computer dynamics and a sufficient sampling of the conformational power several approaches toward the in silico folding space. -

An Ab Initio Materials Simulation Code
CP2K: An ab initio materials simulation code Lianheng Tong Physics on Boat Tutorial , Helsinki, Finland 2015-06-08 Faculty of Natural & Mathematical Sciences Department of Physics Brief Overview • Overview of CP2K - General information about the package - QUICKSTEP: DFT engine • Practical example of using CP2K to generate simulated STM images - Terminal states in AGNR segments with 1H and 2H termination groups What is CP2K? Swiss army knife of molecular simulation www.cp2k.org • Geometry and cell optimisation Energy and • Molecular dynamics (NVE, Force Engine NVT, NPT, Langevin) • STM Images • Sampling energy surfaces (metadynamics) • DFT (LDA, GGA, vdW, • Finding transition states Hybrid) (Nudged Elastic Band) • Quantum Chemistry (MP2, • Path integral molecular RPA) dynamics • Semi-Empirical (DFTB) • Monte Carlo • Classical Force Fields (FIST) • And many more… • Combinations (QM/MM) Development • Freely available, open source, GNU Public License • www.cp2k.org • FORTRAN 95, > 1,000,000 lines of code, very active development (daily commits) • Currently being developed and maintained by community of developers: • Switzerland: Paul Scherrer Institute Switzerland (PSI), Swiss Federal Institute of Technology in Zurich (ETHZ), Universität Zürich (UZH) • USA: IBM Research, Lawrence Livermore National Laboratory (LLNL), Pacific Northwest National Laboratory (PNL) • UK: Edinburgh Parallel Computing Centre (EPCC), King’s College London (KCL), University College London (UCL) • Germany: Ruhr-University Bochum • Others: We welcome contributions from -

Real-Time Pymol Visualization for Rosetta and Pyrosetta
Real-Time PyMOL Visualization for Rosetta and PyRosetta Evan H. Baugh1, Sergey Lyskov1, Brian D. Weitzner1, Jeffrey J. Gray1,2* 1 Department of Chemical and Biomolecular Engineering, The Johns Hopkins University, Baltimore, Maryland, United States of America, 2 Program in Molecular Biophysics, The Johns Hopkins University, Baltimore, Maryland, United States of America Abstract Computational structure prediction and design of proteins and protein-protein complexes have long been inaccessible to those not directly involved in the field. A key missing component has been the ability to visualize the progress of calculations to better understand them. Rosetta is one simulation suite that would benefit from a robust real-time visualization solution. Several tools exist for the sole purpose of visualizing biomolecules; one of the most popular tools, PyMOL (Schro¨dinger), is a powerful, highly extensible, user friendly, and attractive package. Integrating Rosetta and PyMOL directly has many technical and logistical obstacles inhibiting usage. To circumvent these issues, we developed a novel solution based on transmitting biomolecular structure and energy information via UDP sockets. Rosetta and PyMOL run as separate processes, thereby avoiding many technical obstacles while visualizing information on-demand in real-time. When Rosetta detects changes in the structure of a protein, new coordinates are sent over a UDP network socket to a PyMOL instance running a UDP socket listener. PyMOL then interprets and displays the molecule. This implementation also allows remote execution of Rosetta. When combined with PyRosetta, this visualization solution provides an interactive environment for protein structure prediction and design. Citation: Baugh EH, Lyskov S, Weitzner BD, Gray JJ (2011) Real-Time PyMOL Visualization for Rosetta and PyRosetta. -
![Automated Construction of Quantum–Classical Hybrid Models Arxiv:2102.09355V1 [Physics.Chem-Ph] 18 Feb 2021](https://docslib.b-cdn.net/cover/7378/automated-construction-of-quantum-classical-hybrid-models-arxiv-2102-09355v1-physics-chem-ph-18-feb-2021-177378.webp)
Automated Construction of Quantum–Classical Hybrid Models Arxiv:2102.09355V1 [Physics.Chem-Ph] 18 Feb 2021
Automated construction of quantum{classical hybrid models Christoph Brunken and Markus Reiher∗ ETH Z¨urich, Laboratorium f¨urPhysikalische Chemie, Vladimir-Prelog-Weg 2, 8093 Z¨urich, Switzerland February 18, 2021 Abstract We present a protocol for the fully automated construction of quantum mechanical-(QM){ classical hybrid models by extending our previously reported approach on self-parametri- zing system-focused atomistic models (SFAM) [J. Chem. Theory Comput. 2020, 16 (3), 1646{1665]. In this QM/SFAM approach, the size and composition of the QM region is evaluated in an automated manner based on first principles so that the hybrid model describes the atomic forces in the center of the QM region accurately. This entails the au- tomated construction and evaluation of differently sized QM regions with a bearable com- putational overhead that needs to be paid for automated validation procedures. Applying SFAM for the classical part of the model eliminates any dependence on pre-existing pa- rameters due to its system-focused quantum mechanically derived parametrization. Hence, QM/SFAM is capable of delivering a high fidelity and complete automation. Furthermore, since SFAM parameters are generated for the whole system, our ansatz allows for a con- venient re-definition of the QM region during a molecular exploration. For this purpose, a local re-parametrization scheme is introduced, which efficiently generates additional clas- sical parameters on the fly when new covalent bonds are formed (or broken) and moved to the classical region. arXiv:2102.09355v1 [physics.chem-ph] 18 Feb 2021 ∗Corresponding author; e-mail: [email protected] 1 1 Introduction In contrast to most protocols of computational quantum chemistry that consider isolated molecules, chemical processes can take place in a vast variety of complex environments. -

Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development
processes Review Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development Outi M. H. Salo-Ahen 1,2,* , Ida Alanko 1,2, Rajendra Bhadane 1,2 , Alexandre M. J. J. Bonvin 3,* , Rodrigo Vargas Honorato 3, Shakhawath Hossain 4 , André H. Juffer 5 , Aleksei Kabedev 4, Maija Lahtela-Kakkonen 6, Anders Støttrup Larsen 7, Eveline Lescrinier 8 , Parthiban Marimuthu 1,2 , Muhammad Usman Mirza 8 , Ghulam Mustafa 9, Ariane Nunes-Alves 10,11,* , Tatu Pantsar 6,12, Atefeh Saadabadi 1,2 , Kalaimathy Singaravelu 13 and Michiel Vanmeert 8 1 Pharmaceutical Sciences Laboratory (Pharmacy), Åbo Akademi University, Tykistökatu 6 A, Biocity, FI-20520 Turku, Finland; ida.alanko@abo.fi (I.A.); rajendra.bhadane@abo.fi (R.B.); parthiban.marimuthu@abo.fi (P.M.); atefeh.saadabadi@abo.fi (A.S.) 2 Structural Bioinformatics Laboratory (Biochemistry), Åbo Akademi University, Tykistökatu 6 A, Biocity, FI-20520 Turku, Finland 3 Faculty of Science-Chemistry, Bijvoet Center for Biomolecular Research, Utrecht University, 3584 CH Utrecht, The Netherlands; [email protected] 4 Swedish Drug Delivery Forum (SDDF), Department of Pharmacy, Uppsala Biomedical Center, Uppsala University, 751 23 Uppsala, Sweden; [email protected] (S.H.); [email protected] (A.K.) 5 Biocenter Oulu & Faculty of Biochemistry and Molecular Medicine, University of Oulu, Aapistie 7 A, FI-90014 Oulu, Finland; andre.juffer@oulu.fi 6 School of Pharmacy, University of Eastern Finland, FI-70210 Kuopio, Finland; maija.lahtela-kakkonen@uef.fi (M.L.-K.); tatu.pantsar@uef.fi -

Natural Bond Orbital Analysis in the ONETEP Code: Applications to Large Protein Systems Louis P
WWW.C-CHEM.ORG FULL PAPER Natural Bond Orbital Analysis in the ONETEP Code: Applications to Large Protein Systems Louis P. Lee,*[a] Daniel J. Cole,[a] Mike C. Payne,[a] and Chris-Kriton Skylaris[b] First principles electronic structure calculations are typically Generalized Wannier Functions of ONETEP to natural atomic performed in terms of molecular orbitals (or bands), providing a orbitals, NBO analysis can be performed within a localized straightforward theoretical avenue for approximations of region in such a way that ensures the results are identical to an increasing sophistication, but do not usually provide any analysis on the full system. We demonstrate the capabilities of qualitative chemical information about the system. We can this approach by performing illustrative studies of large derive such information via post-processing using natural bond proteins—namely, investigating changes in charge transfer orbital (NBO) analysis, which produces a chemical picture of between the heme group of myoglobin and its ligands with bonding in terms of localized Lewis-type bond and lone pair increasing system size and between a protein and its explicit orbitals that we can use to understand molecular structure and solvent, estimating the contribution of electronic delocalization interactions. We present NBO analysis of large-scale calculations to the stabilization of hydrogen bonds in the binding pocket of with the ONETEP linear-scaling density functional theory package, a drug-receptor complex, and observing, in situ, the n ! p* which we have interfaced with the NBO 5 analysis program. In hyperconjugative interactions between carbonyl groups that ONETEP calculations involving thousands of atoms, one is typically stabilize protein backbones. -

User Manual for the Uppsala Quantum Chemistry Package UQUANTCHEM V.35
User manual for the Uppsala Quantum Chemistry package UQUANTCHEM V.35 by Petros Souvatzis Uppsala 2016 Contents 1 Introduction 6 2 Compiling the code 7 3 What Can be done with UQUANTCHEM 9 3.1 Hartree Fock Calculations . 9 3.2 Configurational Interaction Calculations . 9 3.3 M¨ollerPlesset Calculations (MP2) . 9 3.4 Density Functional Theory Calculations (DFT)) . 9 3.5 Time Dependent Density Functional Theory Calculations (TDDFT)) . 10 3.6 Quantum Montecarlo Calculations . 10 3.7 Born Oppenheimer Molecular Dynamics . 10 4 Setting up a UQANTCHEM calculation 12 4.1 The input files . 12 4.1.1 The INPUTFILE-file . 12 4.1.2 The BASISFILE-file . 13 4.1.3 The BASISFILEAUX-file . 14 4.1.4 The DENSMATSTARTGUESS.dat-file . 14 4.1.5 The MOLDYNRESTART.dat-file . 14 4.1.6 The INITVELO.dat-file . 15 4.1.7 Running Uquantchem . 15 4.2 Input parameters . 15 4.2.1 CORRLEVEL ................................. 15 4.2.2 ADEF ..................................... 15 4.2.3 DOTDFT ................................... 16 4.2.4 NSCCORR ................................... 16 4.2.5 SCERR .................................... 16 4.2.6 MIXTDDFT .................................. 16 4.2.7 EPROFILE .................................. 16 4.2.8 DOABSSPECTRUM ............................... 17 4.2.9 NEPERIOD .................................. 17 4.2.10 EFIELDMAX ................................. 17 4.2.11 EDIR ..................................... 17 4.2.12 FIELDDIR .................................. 18 4.2.13 OMEGA .................................... 18 2 CONTENTS 3 4.2.14 NCHEBGAUSS ................................. 18 4.2.15 RIAPPROX .................................. 18 4.2.16 LIMPRECALC (Only openmp-version) . 19 4.2.17 DIAGDG ................................... 19 4.2.18 NLEBEDEV .................................. 19 4.2.19 MOLDYN ................................... 19 4.2.20 DAMPING ................................... 19 4.2.21 XLBOMD .................................. -

An Explicit-Solvent Conformation Search Method Using Open Software
An explicit-solvent conformation search method using open software Kari Gaalswyk and Christopher N. Rowley Department of Chemistry, Memorial University of Newfoundland, St. John’s, Newfoundland and Labrador, Canada ABSTRACT Computer modeling is a popular tool to identify the most-probable conformers of a molecule. Although the solvent can have a large effect on the stability of a conformation, many popular conformational search methods are only capable of describing molecules in the gas phase or with an implicit solvent model. We have developed a work-flow for performing a conformation search on explicitly-solvated molecules using open source software. This method uses replica exchange molecular dynamics (REMD) to sample the conformational states of the molecule efficiently. Cluster analysis is used to identify the most probable conformations from the simulated trajectory. This work-flow was tested on drug molecules a-amanitin and cabergoline to illustrate its capabilities and effectiveness. The preferred conformations of these molecules in gas phase, implicit solvent, and explicit solvent are significantly different. Subjects Biophysics, Pharmacology, Computational Science Keywords Conformation search, Explicit solvent, Cluster analysis, Replica exchange molecular dynamics INTRODUCTION Many molecules can exist in multiple conformational isomers. Conformational isomers have the same chemical bonds, but differ in their 3D geometry because they hold different torsional angles (Crippen & Havel, 1988). The conformation of a molecule can affect Submitted 5 April 2016 chemical reactivity, molecular binding, and biological activity (Struthers, Rivier & Accepted 6May2016 Published 31 May 2016 Hagler, 1985; Copeland, 2011). Conformations differ in stability because they experience different steric, electrostatic, and solute-solvent interactions. The probability, p,ofa Corresponding author Christopher N. -
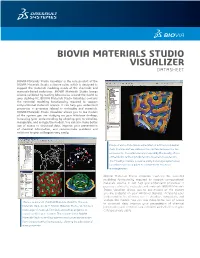
Biovia Materials Studio Visualizer Datasheet
BIOVIA MATERIALS STUDIO VISUALIZER DATASHEET BIOVIA Materials Studio Visualizer is the core product of the BIOVIA Materials Studio software suite, which is designed to support the materials modeling needs of the chemicals and materials-based industries. BIOVIA Materials Studio brings science validated by leading laboratories around the world to your desktop PC. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. You can also make better use of access to structural data, improve your presentation of chemical information, and communicate problems and solutions to your colleagues very easily. Image of early-stage phase segregation in a diblock copolymer melt. The blue surface indicates the interface between the two components. The volume is colormapped by the density of one of the blocks, red being high density, blue being low-density. The MesoDyn module is used to study these large systems over long-times such as required to observe these structural rearrangements. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models.