Maulana Azad Library, Aligarh Muslim University
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
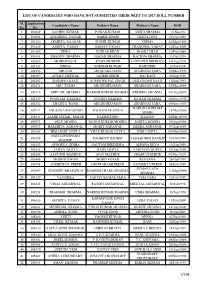
List of Candidates Who Have Not Submitted Their Neet Ug 2017 Roll Number
LIST OF CANDIDATES WHO HAVE NOT SUBMITTED THEIR NEET UG 2017 ROLL NUMBER Sl. Application Candidate's Name Father's Name Mother's Name DOB No. No. 1 100249 SACHIN KUMAR PAWAN KUMAR ANITA SHARMA 15.Nov.93 2 100808 ANUSHEEL NAGAR OMBIR SINGH GEETA DEVI 07/Oct/1998 3 101222 AKSHITA DAAGAR SUSHIL KUMAR SEEMA 24/May/1999 4 101469 ANKITA YADAV SANJAY YADAV CHANCHAL YADAV 31/Dec/1999 5 101593 ZEWA NAWAB KHAN NOOR JAHAN 10/Feb/1999 6 101814 SHAGUN SHARMA GAGAN SHARMA RACHNA SHARMA 13/Oct/1998 7 102087 MOHD FAISAL SHAHABUDDIN JANNATUL FIRDOUS 14/Aug/1998 8 102121 SNEHA SOBODH KUMAR RAJENDRI 20/Jul/1998 9 102256 ABUZAR MD SHAHZAMAN SHABNAM SABA 25/Mar/1999 10 102397 ANJALI DESWAL JAGBIR SINGH RAJ RANI 05/Sep/1998 11 102402 RASMEET KAUR SURINDER PAL SINGH GURVINDER KAUR 11/Sep/1997 12 102474 ABU TALHA MD SHAHZAMAN SHABNAM SABA 25/Mar/1999 13 102515 SHIVANI SHARMA RAKESH KUMAR SHARMA KRISHNA SHARMA 01/Aug/2000 14 102529 POONAM SHARMA GOVIND SHARMA RAJESH SHARMA 04/Nov/1998 15 102532 SHAISTA BANO MD SHAHZAMAN SHABNAM SABA 09/Nov/1997 KARUNA KUMARI 16 102711 ANUSHKA RAJ SINGH RAJ KUMAR SINGH 12/Mar/2000 SINGH 17 102841 AAMIR SUHAIL MALIK NASIMUDDIN SHANNO 26/May/1996 18 102971 ABLE MOGHA MANOJ KUMAR MOGHA SARITA MOGHA 19/Aug/1998 19 103053 HARSHITA AGRAWAL MOHIT AGRAWAL MEERA AGRAWAL 07/Sep/1999 20 103245 HIMANSHI GUPTA VIJAY KUMAR GUPTA INDU GUPTA 09/May/2000 NGULLIENTHANG 21 103428 PAOKHUP HAOKIP VANKHOMOI HAOKIP 03/Oct/1998 HAOKIP 22 103433 APOORVA SINHA GAUTAM BIRENDRA ALPANA DEVA 12/Sep/1999 23 103518 TANYA GUPTA RAJIV GUPTA VANDANA GUPTA 29/Apr/1999 -
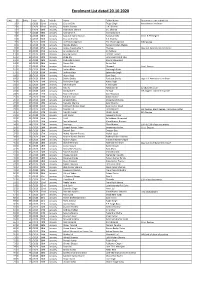
Enrolment List Dated 20.10.2020
Enrolment List dated 20.10.2020 S.No. D/ EnNo Year Date Month Name Father Name Documents to be submitted 1 D/ 1 /2020 02nd January, Gaurav Dixit Tejpal Singh Attendance Certificate 2 D/ 2 /2020 02nd January, Mohit Sharma U.K. Sharma 3 D/ 3 /2020 06th January, Shailendra Sharma S.C. Sharma 4 D/ 4 /2020 06th January, Kasiraman K. Kannuchamy K. 5 D/ 5 /2020 06th January, Saurabh Kumar Suman Ramakant Jha Grad. & PG Degree 6 D/ 6 /2020 06th January, Gaurav Sharma K.K. Sharma 7 D/ 7 /2020 06th January, Jai Prakash Agrawal Tek Chand Agrawal FIR Pending 8 D/ 8 /2020 07th January, Renuka Madan Balram Krishan Madan 9 D/ 9 /2020 07th January, Arokia Arputha Raj T. Thomas Org. LLB. Attendance Certificate 10 D/ 10 /2020 07th January, Anandakumar P. O. Paldurai 11 D/ 11 /2020 08th January, Anurag Kumar Late B.L. Grover 12 D/ 12 /2020 08th January, Atika Alvi Late Umar Mohd. Alvi 13 D/ 13 /2020 08th January, Shahrukh Kirmani Shariq Mahmood 14 D/ 14 /2020 08th January, Shreya Bali Sanjay Bali 15 D/ 15 /2020 09th January, Ashok Kumar Dhanpal Grad. Degree 16 D/ 16 /2020 09th January, Sachin Bhati Mansingh Bhati 17 D/ 17 /2020 09th January, Jaskiran Kaur Satwinder Singh 18 D/ 18 /2020 09th January, Hitain Bajaj Sunil Bajaj 19 D/ 19 /2020 09th January, Shikha Shukla Ravitosh Shukla Org. LLB. Attendace Certificate 20 D/ 20 /2020 10th Janaury, Paramjeet Singh Avtar Singh 21 D/ 21 /2020 10th Janaury, Kalimuthu M. K. Murugan 22 D/ 22 /2020 13th January, Raju N. -

Doctor of Philosophy
Nanoparticle induced suppression of microbial biofilm THESIS SUBMITTED FOR THE AWARD OF THE DEGREE OF Doctor of Philosophy IN BIOTECHNOLOGY BY Shatavari Kulshrestha UNDER THE SUPERVISION OF Dr. ASAD ULLAH KHAN INTERDISCIPLINARY BIOTECHNOLOGY UNIT ALIGARH MUSLIM UNIVERSITY ALIGARH (INDIA) 2015 Certificate This is to certify that the thesis entitled “Nanoparticle induced suppression of microbial biofilm” herewith submitted by Shatavari Kulshrestha, in fulfilment of the requirements for the degree of Doctor of philosophy in Biotechnology of the Aligarh Muslim University, is an authentic record of the research work carried out by her under my supervision and guidance and that no part, thereof, has been presented before for any other degree. Prof. Asad Ullah Khan (Supervisor) Declaration I, hereby, declare that the thesis entitled “Nanoparticle induced suppression of microbial biofilm” is free from any sort of plagiarism. (Shatavari Kulshrestha) Declaration I, hereby, declare that the thesis entitled “Nanoparticle induced suppression of microbial biofilm” embodies the work carried out by me. (Shatavari Kulshrestha) COURSE WORK/COMPREHENSIVE EXAMINATION/ PRE- SUBMISSION SEMINAR COMPLETION CERTIFICATE This is to certify that Ms. Shatavari Kulshrestha, Interdisciplinary Biotechnology Unit, has satisfactorily completed the Course work/Comprehensive examination/Pre-submission seminar requirement which is a part of her Ph.D. programme. Date: ……………… (Signature of the Chairman) Acknowledgement ACKNOWLEDGEMENT All praises to the God, the almighty for providing me this opportunity and granting me the capability to proceed successfully ….. I take this opportunity to express my deep and sincere gratitude to my supervisor Dr. Asad U Khan, Professor, Interdisciplinary biotechnology unit AMU, for giving me the opportunity to do research and providing invaluable guidance throughout this study. -

Original Article Amplification of Meca Gene in Multi-Drug Resistant
Original Article Amplification of mecA gene in multi-drug resistant Staphylococcus aureus strains from hospital personnel Asad U Khan,1 Ayesha Sultan,1 Anju Tyagi,2 Shazia Zahoor,3 Mohd Akram,1 Sukhminderjit Kaur,3 Mohd Shahid,2 Chetana V Vaishnavi 1 1Interdisciplinary Biotechnology Unit, Aligarh Muslim University Aligarh 202002 India; 2Microbiology Department JNMC, AMU, Aligarh, India; 3Gastroenterology Department, PGIMER, Chandigarh, India. Abstract Background: Antibiotic resistance is common among bacterial pathogens associated with both community acquired and nosocomial infections. In view of the present problem of drug resistance we investigated the prevalence of methicillin resistant Staphylococcus aureus (MRSA) and amplified the mecA gene in the isolates from the hand swabs of the hospital personnel. Methodology: The nuc gene was amplified to characterize these isolates at species level. The S. aureus isolates were analyzed for their susceptibility to different classes of antibiotics using the disk diffusion method. The spot inoculation test was performed to detect methicillinase production in these isolates. Results: In the screened isolates of S. aureus , 14.2 and 15 kb of plasmids were present. These isolates showed pronounced resistance against β-lactam antibiotics including second- and third-generation cephalosporins, aminoglycosides, macrolides and fluoroquinolone. Some of the isolates included in this study were resistant to three or more antibiotics. Expression of methicillinase was detected by spot inoculation test, and a few of the isolates were found to produce methicillinase. Moreover, mecA gene was also amplified. Of 17 isolates only 7 showed presence of mecA gene. Conclusion: This study highlights the emerging trend of multiple drug resistance in S. aureus strains isolated from hospital personnel working in a premier hospital in North India. -

Pharmacognosy and Phytotherapy Volume 6 Number 5, June 2014 ISSN 2141-2502
Journal of Pharmacognosy and Phytotherapy Volume 6 Number 5, June 2014 ISSN 2141-2502 ABOUT JPP The Journal of Pharmacognosy and Phytotherapy (JPP) is published monthly (one volume per year) by Academic Journals. The Journal of Pharmacognosy and Phytotherapy (JPP) is an open access journal that provides rapid publication (monthly) of articles in all areas of the subject such as ethnobotany, phytochemistry, ethnopharmacology, zoopharmacognosy, medical anthropology etc. The Journal welcomes the submission of manuscripts that meet the general criteria of significance and scientific excellence. Papers will be published shortly after acceptance. All articles published in JPP are peer-reviewed. Submission of Manuscript Submit manuscripts as e-mail attachment to the Editorial Office at: [email protected]. A manuscript number will be mailed to the corresponding author shortly after submission. The Journal of Pharmacognosy and Phytotherapy (JPP) will only accept manuscripts submitted as e-mail attachments. Please read the Instructions for Authors before submitting your manuscript. The manuscript files should be given the last name of the first author. Editors Dr. (Mrs) Banasri Hazra Dr. Maryam Sarwat Research Scientist (U.G.C.) C/O A.M. Khan, House No. 195 Department of Pharmaceutical Technology Jadavpur University Dr. Yong-Jiang Xu Calcutta - 700032 Saw Swee Hock School of Public Health, India National University of Singapore, Singapore. Prof. Dr. Adeolu Alex Adedapo Dr. Yuanxiong Deng Department of Veterinary Physiology, Dept of Pharmaceutical Science Biochemistry and Pharmacology School of Medicine University of Ibadan, Nigeria Hunan Normal University Tongzipo Road 371, Changsha 410013, Dr. Joana S. Amaral Hunan China Campus de Sta Apolónia, Ap. 1134, 5301-857 Bragança, Portugal Prof.