Original Article Amplification of Meca Gene in Multi-Drug Resistant
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Detection of the Meca Gene and Identification of Staphylococcus
braz j infect dis 2 0 1 8;2 2(2):99–105 The Brazilian Journal of INFECTIOUS DISEASES www.elsevier.com/locate/bjid Original article Detection of the mecA gene and identification of Staphylococcus directly from blood culture bottles by multiplex polymerase chain reaction a,b,∗ a Taisa Trevizani Rocchetti , Katheryne Benini Martins , a c Patricia Yoshida Faccioli Martins , Rogério Antonio de Oliveira , d b Alessandro Lia Mondelli , Carlos Magno Castelo Branco Fortaleza , a Maria de Lourdes Ribeiro de Souza da Cunha a UNESP - Univ Estadual Paulista, Instituto de Biociências de Botucatu, Departamento de Microbiologia e Imunologia, Botucatu, SP, Brazil b UNESP - Univ Estadual Paulista, Faculdade de Medicina de Botucatu, Hospital Universitário, Departamento de Doenc¸as Tropicais, Botucatu, SP, Brazil c UNESP - Univ Estadual Paulista, Instituto de Biociências de Botucatu, Departamento de Biociência, Botucatu, SP, Brazil d UNESP - Univ Estadual Paulista, Faculdade de Medicina de Botucatu, Hospital Universitário, Departamento de Medicina Interna, Botucatu, SP, Brazil a r t i c l e i n f o a b s t r a c t Article history: Introduction: Staphylococcus spp. – both S. aureus, including methicillin-resistant strains Received 3 November 2017 (MRSA) and coagulase negative staphylococci (CoNS) – are relevant agents of healthcare- Accepted 18 February 2018 associated infections. Therefore, the rapid recognition of MRSA and methicillin-resistant Available online 13 March 2018 CoNS from blood stream infections is critically important for patient management. It is worth noting that inappropriate empiric therapy has been associated with higher in-hospital Keywords: mortality. Material and methods: In this study we evaluated a multiplex polymerase chain reaction (mul- Staphylococcus spp. -
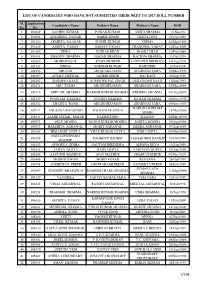
List of Candidates Who Have Not Submitted Their Neet Ug 2017 Roll Number
LIST OF CANDIDATES WHO HAVE NOT SUBMITTED THEIR NEET UG 2017 ROLL NUMBER Sl. Application Candidate's Name Father's Name Mother's Name DOB No. No. 1 100249 SACHIN KUMAR PAWAN KUMAR ANITA SHARMA 15.Nov.93 2 100808 ANUSHEEL NAGAR OMBIR SINGH GEETA DEVI 07/Oct/1998 3 101222 AKSHITA DAAGAR SUSHIL KUMAR SEEMA 24/May/1999 4 101469 ANKITA YADAV SANJAY YADAV CHANCHAL YADAV 31/Dec/1999 5 101593 ZEWA NAWAB KHAN NOOR JAHAN 10/Feb/1999 6 101814 SHAGUN SHARMA GAGAN SHARMA RACHNA SHARMA 13/Oct/1998 7 102087 MOHD FAISAL SHAHABUDDIN JANNATUL FIRDOUS 14/Aug/1998 8 102121 SNEHA SOBODH KUMAR RAJENDRI 20/Jul/1998 9 102256 ABUZAR MD SHAHZAMAN SHABNAM SABA 25/Mar/1999 10 102397 ANJALI DESWAL JAGBIR SINGH RAJ RANI 05/Sep/1998 11 102402 RASMEET KAUR SURINDER PAL SINGH GURVINDER KAUR 11/Sep/1997 12 102474 ABU TALHA MD SHAHZAMAN SHABNAM SABA 25/Mar/1999 13 102515 SHIVANI SHARMA RAKESH KUMAR SHARMA KRISHNA SHARMA 01/Aug/2000 14 102529 POONAM SHARMA GOVIND SHARMA RAJESH SHARMA 04/Nov/1998 15 102532 SHAISTA BANO MD SHAHZAMAN SHABNAM SABA 09/Nov/1997 KARUNA KUMARI 16 102711 ANUSHKA RAJ SINGH RAJ KUMAR SINGH 12/Mar/2000 SINGH 17 102841 AAMIR SUHAIL MALIK NASIMUDDIN SHANNO 26/May/1996 18 102971 ABLE MOGHA MANOJ KUMAR MOGHA SARITA MOGHA 19/Aug/1998 19 103053 HARSHITA AGRAWAL MOHIT AGRAWAL MEERA AGRAWAL 07/Sep/1999 20 103245 HIMANSHI GUPTA VIJAY KUMAR GUPTA INDU GUPTA 09/May/2000 NGULLIENTHANG 21 103428 PAOKHUP HAOKIP VANKHOMOI HAOKIP 03/Oct/1998 HAOKIP 22 103433 APOORVA SINHA GAUTAM BIRENDRA ALPANA DEVA 12/Sep/1999 23 103518 TANYA GUPTA RAJIV GUPTA VANDANA GUPTA 29/Apr/1999 -

Emergence of Oxacillin-Resistant Staphylococcus Lugdunensis
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Journal of Microbiology, Immunology and Infection (2016) 49, 885e891 Available online at www.sciencedirect.com ScienceDirect journal homepage: www.e-jmii.com ORIGINAL ARTICLE Emergence of oxacillin-resistant Staphylococcus lugdunensis carrying staphylococcal cassette chromosome mec type V in central Taiwan Ting-Yu Yen a,b,g, Yun-Ju Sung c,g, Hsiao-Chuan Lin a,b, Ching-Tien Peng a,b, Ni Tien c, Kao-Pin Hwang a,b, Jang-Jih Lu d,e,f,* a Department of Pediatrics, Children’s Hospital, China Medical University Hospital, Taichung, Taiwan b School of Medicine, China Medical University, Taichung, Taiwan c Department of Laboratory Medicine, China Medical University Hospital, Taichung, Taiwan d Department of Medical Research, China Medical University Hospital, Taichung, Taiwan e Department of Laboratory Medicine, Chang Gung Memorial Hospital at Linkou, Taoyuan, Taiwan f Department of Medical Biotechnology and Laboratory Science, Chang Gung University, Kwei-Shan, Taoyuan, Taiwan Received 5 August 2014; received in revised form 12 November 2014; accepted 29 November 2014 Available online 11 December 2014 KEYWORDS Background: Staphylococcus lugdunensis has emerged as a key pathogen for clinical infection. coagulase-negative It is sensitive to most antistaphylococcal agents, but it is increasingly resistant to b-lactam an- staphylococcus; tibiotics. Oxacillin-resistant S. lugdunensis isolates carrying the mecA gene pose a major oxacillin resistance; concern for therapy failure. SCCmec gene; Methods: To assess the epidemiology and presence of mecA in S. lugdunensis, we gauged the Staphylococcus prevalence and antibiotic resistance of S. -
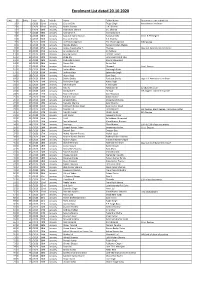
Enrolment List Dated 20.10.2020
Enrolment List dated 20.10.2020 S.No. D/ EnNo Year Date Month Name Father Name Documents to be submitted 1 D/ 1 /2020 02nd January, Gaurav Dixit Tejpal Singh Attendance Certificate 2 D/ 2 /2020 02nd January, Mohit Sharma U.K. Sharma 3 D/ 3 /2020 06th January, Shailendra Sharma S.C. Sharma 4 D/ 4 /2020 06th January, Kasiraman K. Kannuchamy K. 5 D/ 5 /2020 06th January, Saurabh Kumar Suman Ramakant Jha Grad. & PG Degree 6 D/ 6 /2020 06th January, Gaurav Sharma K.K. Sharma 7 D/ 7 /2020 06th January, Jai Prakash Agrawal Tek Chand Agrawal FIR Pending 8 D/ 8 /2020 07th January, Renuka Madan Balram Krishan Madan 9 D/ 9 /2020 07th January, Arokia Arputha Raj T. Thomas Org. LLB. Attendance Certificate 10 D/ 10 /2020 07th January, Anandakumar P. O. Paldurai 11 D/ 11 /2020 08th January, Anurag Kumar Late B.L. Grover 12 D/ 12 /2020 08th January, Atika Alvi Late Umar Mohd. Alvi 13 D/ 13 /2020 08th January, Shahrukh Kirmani Shariq Mahmood 14 D/ 14 /2020 08th January, Shreya Bali Sanjay Bali 15 D/ 15 /2020 09th January, Ashok Kumar Dhanpal Grad. Degree 16 D/ 16 /2020 09th January, Sachin Bhati Mansingh Bhati 17 D/ 17 /2020 09th January, Jaskiran Kaur Satwinder Singh 18 D/ 18 /2020 09th January, Hitain Bajaj Sunil Bajaj 19 D/ 19 /2020 09th January, Shikha Shukla Ravitosh Shukla Org. LLB. Attendace Certificate 20 D/ 20 /2020 10th Janaury, Paramjeet Singh Avtar Singh 21 D/ 21 /2020 10th Janaury, Kalimuthu M. K. Murugan 22 D/ 22 /2020 13th January, Raju N. -

CLONING and TRANSFORMATION of Meca GENE of STAPHYLOCOCCUS AUREUS ISOLATED from HOSPITAL SAMPLES H
Int. J. Chem. Sci.: 11(1), 2013, 518-528 ISSN 0972-768X www.sadgurupublications.com CLONING AND TRANSFORMATION OF MecA GENE OF STAPHYLOCOCCUS AUREUS ISOLATED FROM HOSPITAL SAMPLES H. S. RAVIKUMAR PATIL* T. VASANTHA NAIKa, B. R. VIJAY AVINb and H. A. SAYESWARAc Department of Biotechnology, G. M. Institute of Technology, DAVANGERE – 577006 (K.S.) INDIA aDepartment of Botany, D. R. M. Science College, Davangere University, DAVANGERE – 577006 (K.S.) INDIA bDepartment of P.G. Studies and Research in Biotechnology, Sahyadri Science College (Autonomous), Kuvempu University, SHIVAMOGGA – 577203 (K.S.) INDIA cDepartment of Zoology, Sahyadri Science College (Autonomous), Kuvempu University, SHIVAMOGGA – 577203 (K.S.) INDIA ABSTRACT Antibiotic resistance is common among bacterial pathogens associated with both community acquired and nosocomial infections. In view of the present problem of drug resistance, we investigated the prevalence of methicillin resistant Staphylococcus aureus (MRSA) and amplified the mecA gene in the isolates from the hospital samples. The S. aureus isolates were analyzed for their susceptibility to different classes of antibiotics using Gentamycin and it shows resistance to Kanamycin, Methicillin, Amphotericin and Tetracyclin. MIC test was conducted for S. aureus for antibiotics Ampicillin and Gentamycin. Different samples show different MIC of antibiotics. Plasmid DNA was isolated from the sample 1 and mecA gene was amplified by suitable primers by PCR. This amplified product was sequenced and sequenced size was determined to be 1320 bp. Further it was cloned to plasmid pUC18 vector and transformed to DH5α strains of E. coli. This study highlights the emerging trend of multiple drug resistance in S. aureus samples isolated from hospitals. -

Doctor of Philosophy
Nanoparticle induced suppression of microbial biofilm THESIS SUBMITTED FOR THE AWARD OF THE DEGREE OF Doctor of Philosophy IN BIOTECHNOLOGY BY Shatavari Kulshrestha UNDER THE SUPERVISION OF Dr. ASAD ULLAH KHAN INTERDISCIPLINARY BIOTECHNOLOGY UNIT ALIGARH MUSLIM UNIVERSITY ALIGARH (INDIA) 2015 Certificate This is to certify that the thesis entitled “Nanoparticle induced suppression of microbial biofilm” herewith submitted by Shatavari Kulshrestha, in fulfilment of the requirements for the degree of Doctor of philosophy in Biotechnology of the Aligarh Muslim University, is an authentic record of the research work carried out by her under my supervision and guidance and that no part, thereof, has been presented before for any other degree. Prof. Asad Ullah Khan (Supervisor) Declaration I, hereby, declare that the thesis entitled “Nanoparticle induced suppression of microbial biofilm” is free from any sort of plagiarism. (Shatavari Kulshrestha) Declaration I, hereby, declare that the thesis entitled “Nanoparticle induced suppression of microbial biofilm” embodies the work carried out by me. (Shatavari Kulshrestha) COURSE WORK/COMPREHENSIVE EXAMINATION/ PRE- SUBMISSION SEMINAR COMPLETION CERTIFICATE This is to certify that Ms. Shatavari Kulshrestha, Interdisciplinary Biotechnology Unit, has satisfactorily completed the Course work/Comprehensive examination/Pre-submission seminar requirement which is a part of her Ph.D. programme. Date: ……………… (Signature of the Chairman) Acknowledgement ACKNOWLEDGEMENT All praises to the God, the almighty for providing me this opportunity and granting me the capability to proceed successfully ….. I take this opportunity to express my deep and sincere gratitude to my supervisor Dr. Asad U Khan, Professor, Interdisciplinary biotechnology unit AMU, for giving me the opportunity to do research and providing invaluable guidance throughout this study. -

Methicillin-Resistant Staphylococcus Aureus Meca (Penicillin Binding Protein 2) & S
Techne ® qPCR test Methicillin-resistant Staphylococcus aureus mecA (penicillin binding protein 2) & S. aureus FEMB gene (chromosomal gene) 150 tests For general laboratory and research use only Quantification of Methicillin-resistant Staphylococcus aureus genomes. 1 Advanced kit handbook HB10.07.07 Introduction to Methicillin-resistant Staphylococcus aureus Methicillin-resistant Staphylococcus aureus (MRSA) is a specific strain of the Staphylococcus aureus bacterium that has developed antibiotic resistance to all penicillins, including methicillin and other narrow-spectrum β-lactamase-resistant penicillin antibiotics. [1] The resistant strain, MRSA was first discovered in the UK in 1961 and is now widespread, particularly in the hospital setting where it is commonly termed a superbug. MRSA may also be known as oxacillin-resistant Staphylococcus aureus (ORSA) and multiple-resistant Staphylococcus aureus, while non-methicillin resistant strains of S. aureus are sometimes called methicillin-susceptible Staphylococcus aureus (MSSA) if an explicit distinction must be made. Although MRSA has traditionally been seen as a hospital-associated infection, community- acquired MRSA strains have appeared in recent years, notably in the US and Australia.[2] The abbreviations CA-MRSA (community-associated MRSA) and HA-MRSA (hospital- associated MRSA) are now commonly seen in medical literature. Methicillin resistance arises by acquisition of a staphylococcal cassette chromosome SCCmec, and is conferred by the mecA gene. Expression of this gene yields PBP2a, a penicillin binding protein with reduced affinity for β-lactam rings (the primary active-site of the β-lactam antibiotics).[6] Some strains of S. aureus over-express β-lactamase and appear to be resistant to oxacillin and, rarely, methicillin despite being mecA-negative. -

Detection of Antimicrobial Resistance of Bacteria Staphylococcus Chromogenes Isolated from Sheep’S Milk and Cheese
antibiotics Article Detection of Antimicrobial Resistance of Bacteria Staphylococcus chromogenes Isolated from Sheep’s Milk and Cheese Ivana Regecová 1 , Jana Výrostková 1,* , František Zigo 2 , Gabriela Gregová 3 and Mariana Kováˇcová 1 1 Department of Food Hygiene, Technology and Safety, The University of Veterinary Medicine and Pharmacy in Košice, Komenského 73, 041 81 Košice, Slovakia; [email protected] (I.R.); [email protected] (M.K.) 2 Department of Animal Nutrition and Husbandry, The University of Veterinary Medicine and Pharmacy in Košice, Komenského 73, 041 81 Košice, Slovakia; [email protected] 3 Department of Public Veterinary Medicine and Animal Welfare, The University of Veterinary Medicine and Pharmacy in Košice, Komenského 73, 041 81 Košice, Slovakia; [email protected] * Correspondence: [email protected] Abstract: Antimicrobial and multidrug resistance is detected in nonaureus staphylococci, including Staphylococcus chromogenes, which commonly causes intramammary infections. Recent clinical studies point to the presence of methicillin-resistant S. chromogenes. Therefore, this study aims to determine the prevalence of this species in samples of sheep‘s milk and cheeses made from them. Isolates were identified by polymerase chain reaction and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI–TOF). A total of 208 staphylococcal isolates were identified. Of these, 18% were identified as S. chromogenes. The antimicrobial resistance of the identified isolates was Citation: Regecová, I.; Výrostková, J.; determined using the agar dilution method against penicillin, ceftaroline, teicoplanin, gentamicin, Zigo, F.; Gregová, G.; Kováˇcová,M. Detection of Antimicrobial Resistance erythromycin, tetracycline, and ofloxacin. The highest resistance was found to penicillin (95%), of Bacteria Staphylococcus chromogenes tetracycline (86%), and oxacillin (81%). -

MRSA) to Methicillin Susceptible Staphylococcus Aureus (MSSA
Bitrus et al. BMC Microbiology (2017) 17:83 DOI 10.1186/s12866-017-0994-6 RESEARCH ARTICLE Open Access In vitro transfer of methicillin resistance determinants mecA from methicillin resistant Staphylococcus aureus (MRSA) to methicillin susceptible Staphylococcus aureus (MSSA) Asinamai Athliamai Bitrus1, Zakaria Zunita1*, Siti Khairani Bejo1, Sarah Othman2 and Nur Adilah Ahmad Nadzir1 Abstract Background: Staphylococcus aureus more than any other human pathogen is a better model for the study of the adaptive evolution of bacterial resistance to antibiotics, as it has demonstrated a remarkable ability in its response to new antibiotics. This study was designed to investigate the in vitro transfer of mecA gene from methicillin resistant S. aureus to methicillin susceptible S. aureus. Result: The recipient transconjugants were resistant to erythromycin, cefpodoxime and were mecA positive. PCR amplification of mecA after mix culture plating on Luria Bertani agar containing 100 μg/mL showed that 75% of the donor and 58.3% of the recipient transconjugants were mecA positive. Additionally, 61.5% of both the donor cells and recipient transconjugants were mecA positive, while 46.2% and 41.75% of both donor and recipient transconjugants were mecA positive on LB agar containing 50 μg/mL and 30 μg/mL respectively. Conclusion: In this study, the direction of transfer of phenotypic resistance as well as mecA was observed to have occurred from the donor to the recipient strains. This study affirmed the importance of horizontal transfer events in the dissemination of antibiotics resistance among different strains of MRSA. Keywords: Antibiotics, Horizontal gene transfer, Methicillin, Resistance, Staphylococcus aureus Background fields of veterinary and human medicine as well as in Staphylococcus aureus is a good model better than any animal husbandry [1]. -

Significance of MRSA in Nosocomial Infections
International Journal of Applied Science www.ijas.org.uk Review Article Significance of MRSA in nosocomial Infections Dr. Javid Ahmad Bhat*1 and Dr Rajesh Tenguria2 1Division of Microbiology, M.V.M College Bhopal, M.P. India 2Department of Microbiology, M.V.M. College Bhopal, M.P. India. A R T I C L E I N F O A B S T R A C T Received 25 June 2014 Received in revised form 29 June 2014 Staphylococcus aureus is one of the most virulent microbial pathogen Accepted 04 July 2014 amongst gram positive bacteria to cause nosocomial and community acquired infections. An additional concern is the emergence and Keywords: dissemination of nosocomial organisms with increased resistance to MRSA, antimicrobial agents. Such microbes include methicillin resistant VRE, Nosocomial, S.aureus (MRSA), S.epidermidis, vancomycin resistant Enterococci mecA gene, (VRE) and VISA. The development of vaccines and drugs that prevent CA-MRSA. and cure bacterial infections was one of the twentieth century’s major contributions to human longevity and quality of life. Antibacterial agents are amongst the most commonly prescribed drugs of any kind worldwide. Used appropriately, these drugs are lifesaving however, their indiscriminate use drives up the cost of health care leading to a plethora of side effects and drug interactions and fosters the emergence of bacterial resistance rendering previously valuable drugs useless. Corresponding author: Division of Microbiology, M.V.M College Bhopal, M.P. India © 2014 International Journal of Applied Science- All rights reserved E-mail address: [email protected] INTRODUCTION Staphylococcus aureus is one of the Staphylococcus aureus is universal in most common and important pathogen, distribution found in pus, boils, abscess, skin, responsible for the majority of nosocomial throat, nasophrynx, oral mucosa, soil, sewage, infections. -

Detection of Resistance Meca Gene in Gram Positive Bacteria Described As Nosocomial
American Journal of www.biomedgrid.com Biomedical Science & Research ISSN: 2642-1747 --------------------------------------------------------------------------------------------------------------------------------- Research Article Copy Right@ Navarro C Detection of Resistance mecA Gene In Gram Positive Bacteria Described as Nosocomial Zúñiga B1, Jara MA1, Mosnaim AD2 and Navarro C1* 1Department of Preventive Animal Medicine, University of Chile, Chile 2Cellular and Molecular Pharmacology, Rosalind Franklin University of Medicine and Science, USA *Corresponding author: Navarro C, Department of Preventive Animal Medicine, University of Chile, Chile To Cite This Article: Navarro C. Detection of Resistance mecA Gene In Gram Positive Bacteria Described as Nosocomial. Am J Biomed Sci & Res. 2018 - 4(5). AJBSR.MS.ID.000831. DOI: 10.34297/AJBSR.2019.04.000831 Received: August 08, 2019; Published: August 21, 2019 Abstract is natural in some bacteria, while in others it is a condition acquired through the incorporation of genes that code various mechanisms of resistance. Shortly after the introduction of an antimicrobial to the market it has been possible to find bacteria that are resistant to its action. This resistance These resistant strains represent a big problem in human hospitals when causing nosocomial infections, since the therapeutic options are limited. In veterinary medicine, although nosocomial infections are increasing, they are still less studied than those acquired by people. However, these infections -both in human and veterinary patients- have in common to be caused, mainly, by methicillin-resistant staphylococci. Due to this fact, in addition to the fact that methicillin-resistant Staphylococcus aureus is transmitted between different animal species, including humans, the purpose of this work was to detect the mecA resistance gene in bacteria described as nosocomial. -

Study of Methicillin Beta Lactam Antibiotic with Penicillin Binding Protein 2A B
B. Padmavathi Bai RJLBPCS 2019 www.rjlbpcs.com Life Science Informatics Publications Original Research Article DOI: 10.26479/2019.0502.47 STUDY OF METHICILLIN BETA LACTAM ANTIBIOTIC WITH PENICILLIN BINDING PROTEIN 2A B. Padmavathi Bai Department of Botany, S.N.S.R Degree college, Velgode, Kurnool, India. ABSTRACT: The mecA gene is a gene found in bacterial cells. The mecA gene allows a bacterium to be resistant to antibiotics such as methicillin, penicillin and other penicillin-like antibiotics. The most commonly known carrier of the mecA gene is the bacterium known as MRSA. Apart from Staphylococcus aureus and other Staphylococcus species, it can also be found in Streptococcus pneumoniae strains resistant to penicillin-like antibiotics. In Staphylococcus species, mecA is spread on the SCCmec genetic element. The mecA gene does not allow the ring like structure of penicillin- like antibiotics to bind to the enzymes that help form the cell wall of the bacterium (transpeptidases), and hence the bacteria is able to replicate as normal. The gene encodes the protein PBP2A (penicillin binding protein 2A). PBP2A has a low affinity for beta-lactam antibiotics such as methicillin and penicillin. This enables transpeptidase activity in the presence of beta-lactams, preventing them from inhibiting cell wall synthesis. KEYWORDS: Methicillin, Mec A gene, Staphylococcus aureus, PBP2A, Beta lactam. Corresponding Author: Dr. B. Padmavathi Bai* Ph.D. Department of Botany, S.N.S.R Degree college, Velgode, Kurnool, India. 1.INTRODUCTION The multiple antibiotic resistances of methicillin-resistant strains of Staphylococcus aureus (MRSA) has become a major clinical problem worldwide. Rates of MRSA infection are increasing [1].