122220 APP 500+ Pesticides Fruit Veggies4.Indd
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Historical Perspectives on Apple Production: Fruit Tree Pest Management, Regulation and New Insecticidal Chemistries
Historical Perspectives on Apple Production: Fruit Tree Pest Management, Regulation and New Insecticidal Chemistries. Peter Jentsch Extension Associate Department of Entomology Cornell University's Hudson Valley Lab 3357 Rt. 9W; PO box 727 Highland, NY 12528 email: [email protected] Phone 845-691-7151 Mobile: 845-417-7465 http://www.nysaes.cornell.edu/ent/faculty/jentsch/ 2 Historical Perspectives on Fruit Production: Fruit Tree Pest Management, Regulation and New Chemistries. by Peter Jentsch I. Historical Use of Pesticides in Apple Production Overview of Apple Production and Pest Management Prior to 1940 Synthetic Pesticide Development and Use II. Influences Changing the Pest Management Profile in Apple Production Chemical Residues in Early Insect Management Historical Chemical Regulation Recent Regulation Developments Changing Pest Management Food Quality Protection Act of 1996 The Science Behind The Methodology Pesticide Revisions – Requirements For New Registrations III. Resistance of Insect Pests to Insecticides Resistance Pest Management Strategies IV. Reduced Risk Chemistries: New Modes of Action and the Insecticide Treadmill Fermentation Microbial Products Bt’s, Abamectins, Spinosads Juvenile Hormone Analogs Formamidines, Juvenile Hormone Analogs And Mimics Insect Growth Regulators Azadirachtin, Thiadiazine Neonicotinyls Major Reduced Risk Materials: Carboxamides, Carboxylic Acid Esters, Granulosis Viruses, Diphenyloxazolines, Insecticidal Soaps, Benzoyl Urea Growth Regulators, Tetronic Acids, Oxadiazenes , Particle Films, Phenoxypyrazoles, Pyridazinones, Spinosads, Tetrazines , Organotins, Quinolines. 3 I Historical Use of Pesticides in Apple Production Overview of Apple Production and Pest Management Prior to 1940 The apple has a rather ominous origin. Its inception is framed in the biblical text regarding the genesis of mankind. The backdrop appears to be the turbulent setting of what many scholars believe to be present day Iraq. -

Mode of Action Classification
Gr oup 15: Inhibitors of chitin biosynthe si s, type 0 (Only m ajor r epr esentatives of the gr oup ar e shown) Gr oup 1: Acetylcholi ne st er a se (AChE) inhibitors (Only major representatives of the groups are shown) Mode of Action Classification Flufenoxuron No v a l u ro n Triflumuron Ac ephate Me t h a mi d o p h o s Aldic arb Ox a my l Di fl u b e n z u ron Lufenuron Teflubenzuron 15 Benz oy lur eas Me t h i o c a r b Fenitrothion Gr oup 16: Inhibitors of Gr oup 17: Moulting Gr oup 18: Ecdysone r eceptor agonists Thiodicarb Ch l o rp y ri fo s Ca rb a ry l Profenofos chitin biosynthesis, disr uptor , D ipter an type 1 Me t h o my l Ma l a t h i o n Insecticide Resistance Action Committee Pirimic arb Di meth o a te Ch ro mafe n o z i d e Ca rb o fu ra n Triazophos Me t h o x y f e n o z id e 1A Carbamates 1B Organophosphates The Key to Resistance Management Buprofez in Cy ro maz i n e 18 Diacyl- Ha l o fe n o z i d e Tebufenozide Ø Successive generations of a pest should not be treated with compounds from the same MoA Gr oup. 16 Buprofezin 17 Cyromazine hydrazines Gr oup 2: GABA-gated chlor ide channel antagonis ts Ø Not all of the cur r ent gr oupings ar e based on knowledg e of a shar ed tar get pr otein. -

Neuroactive Insecticides: Targets, Selectivity, Resistance, and Secondary Effects
EN58CH06-Casida ARI 5 December 2012 8:11 Neuroactive Insecticides: Targets, Selectivity, Resistance, and Secondary Effects John E. Casida1,∗ and Kathleen A. Durkin2 1Environmental Chemistry and Toxicology Laboratory, Department of Environmental Science, Policy, and Management, 2Molecular Graphics and Computational Facility, College of Chemistry, University of California, Berkeley, California 94720; email: [email protected], [email protected] Annu. Rev. Entomol. 2013. 58:99–117 Keywords The Annual Review of Entomology is online at acetylcholinesterase, calcium channels, GABAA receptor, nicotinic ento.annualreviews.org receptor, secondary targets, sodium channel This article’s doi: 10.1146/annurev-ento-120811-153645 Abstract Copyright c 2013 by Annual Reviews. Neuroactive insecticides are the principal means of protecting crops, people, All rights reserved livestock, and pets from pest insect attack and disease transmission. Cur- ∗ Corresponding author rently, the four major nerve targets are acetylcholinesterase for organophos- phates and methylcarbamates, the nicotinic acetylcholine receptor for neonicotinoids, the γ-aminobutyric acid receptor/chloride channel for by Public Health Information Access Project on 04/29/14. For personal use only. Annu. Rev. Entomol. 2013.58:99-117. Downloaded from www.annualreviews.org polychlorocyclohexanes and fiproles, and the voltage-gated sodium channel for pyrethroids and dichlorodiphenyltrichloroethane. Species selectivity and acquired resistance are attributable in part to structural differences in binding subsites, receptor subunit interfaces, or transmembrane regions. Additional targets are sites in the sodium channel (indoxacarb and metaflumizone), the glutamate-gated chloride channel (avermectins), the octopamine receptor (amitraz metabolite), and the calcium-activated calcium channel (diamides). Secondary toxic effects in mammals from off-target serine hydrolase inhibi- tion include organophosphate-induced delayed neuropathy and disruption of the cannabinoid system. -

The Pesticides Are Generally Classified Into Various Groups Based
Lecture 19 PESTICIDES GROUPS Groups of pesticides : The pesticides are generally classified into various groups based on pest organism against which the compounds are used, their chemical nature, mode of entry and mode of action. 1. Based on organisms a) Insecticides : Chemicals used to kill or control insects (eg.) endosulfan, malathion b) Rodenticides : Chemicals exclusively used to control rats (eg.) Zinc phosphide c) Acaricides : Chemicals used to control mites on crops / animals (eg.) Dicofol d) Avicides : Chemicals used to repel the birds (eg.) Anthraquionone e) Molluscicides : Chemicals used to kill the snails and slugs (eg.) Metaldehyde f) Nematicides : Chemicals used to control nematodes (eg.) Ethylene dibromide g) Fungicides : Chemicals used to control plant diseases caused by fungi (eg.) Copper oxy cholirde h) Bactericide : Chemicals used to control the plant diseases caused by bacteria (eg.) Streptomycin sulphate i) Herbicide : Chemicals used to control weeds (eg.) 2,4, - D 2. Based on mode of entry a) Stomach poison : The insecticide applied in the leaves and other parts of the plant when ingested, act in the digestive system of the insect and bring about kill (eg.) Malathion. b) Contact Poison : The toxicant which brings about death of the pest species by means of contact (eg.) Fenvalerate. c) Fumigant : Toxicant enter in vapour form into the tracheal system (respiratory poison) through spiracles (eg.) Aluminium phosphide d) Systemic poison : Chemicals when applied to plant or soil are absorbed by foliage (or) roots and translocated through vascular system and cause death of insect feeding on plant. (eg.) Dimethoate. 3. Based on mode of action a) Physical poison : Toxicant which brings about kill of one insect by exerting a physical effect (eg.) Activated clay. -

Hydrazone Compounds, Processes for Their Production, Intermediates Useful for Their Production and Pesticidal Compositions Containing Them
Europaisches Patentamt J European Patent Office © Publication number: 0 500 111 A2 Office europeen des brevets © EUROPEAN PATENT APPLICATION © Application number: 92102869.2 © int. CIA C07C 251/88, A01N 35/10, C07C 257/22, A01 N 37/52, @ Date of filing: 20.02.92 C07C 309/66, C07C 257/06, C07C 281/04, C07C 49/76, C07C 49/80, C07C 49/807 ® Priority: 22.02.91 JP 114191/91 Sangyo Kaisha Ltd Chuo Kenkyusho, 3-1, Nishi-shibukawa @ Date of publication of application: 2-chome 26.08.92 Bulletin 92/35 Kusatsu-shi, Shiga-ken(JP) Inventor: Sasaki, Hiroshi, c/o Ishihara Sangyo © Designated Contracting States: Kaisha Ltd. AT BE CH DE DK ES FR GB GR IT LI LU MC Chuo Kenkyusho, 3-1, Nishi-shibukawa NL PT SE 2-chome Kusatsu-shi, Shiga-ken(JP) © Applicant: ISHIHARA SANGYO KAISHA, LTD. Inventor: Morita, Masayuki, c/o Ishihara No. 3-22, Edobori 1-chome Sangyo Kaisha Ltd. Nishi-ku Osaka(JP) Chuo Kenkyusho, 3-1, Nishi-shibukawa 2-chome @ Inventor: Toki, Tadaaki, c/o Ishihara Sangyo Kusatsu-shi, Shiga-ken(JP) Kaisha Ltd. Inventor: Yoneda, Tetsuo, c/o Ishihara Chuo Kenkyusho, 3-1, Nishi-shibukawa Sangyo Kaisha Ltd. 2-chome Chuo Kenkyusho, 3-1, Nishi-shibukawa Kusatsu-shi, Shiga-ken(JP) 2-chome Inventor: Koyanagi, Toru, c/o Ishihara Sangyo Kusatsu-shi, Shiga-ken(JP) Kaisha Ltd. Chuo Kenkyusho, 3-1, Nishi-shibukawa 2-chome © Representative: Wachtershauser, Gunter, Dr. Kusatsu-shi, Shiga-ken(JP) Tal 29 Inventor: Yoshida, Kiyomitsu, c/o Ishihara W-8000 Munchen 2(DE) © Hydrazone compounds, processes for their production, intermediates useful for their production and pesticidal compositions containing them. -

Pesticide Resistance in Bed Bugs Everywhere!!!!!
2/24/2018 Pesticide Resistance in Bed bugs were virtually eradicated from the U.S. in Bed Bugs the post WWII era due to DDT and other powerful Shujuan (Lucy) Li insecticides. University of Arizona Alvaro Romero New Mexico State University 2 By the 1960s, bed bugs had developed resistance Public housing Apartments to DDT, methoxychlor and analogues, BHC, Schools dieldrin and analogues , and pyrethrins ( Busvine 1958, Hospitals Nursing homes Cwilich & Mer 1957, Mallis and Miller 1964 ) . Homes Transportation Child care Medical facilities Hotels & motels Health care facilities Airports Movie theaters Department stores Products, vendors, or commercial services mentioned or pictured in this seminar are for Everywhere!!!!! illustrative purposes only and are not meant to be endorsements. 3 4 University of Arizona; Arizona Pest Management Center 1 2/24/2018 Possible reasons for treatment failure? Missed some Clutter Reintroduction Have you seen these after treatments? 5 6 Dose - response assays for field - collected strains Bed bugs survived direct insecticide sprays 99 deltamethrin 90 Ft. Dix F1 50 ) e l a c 10 s t CIN1 i b o 1.0 r p ( y t i l a t r 99 - cyhalothrin o m e 90 g a t n Resistance ratio (RR) at least 6,000 !!! e c Ft. Dix r 50 e P 10 CIN1 Suspend® ( Deltamethrin ) 1.0 10 -7 10 -6 10 -5 10 -4 10 -3 10 -2 10 -1 10 0 10 1 10 2 10 3 10 4 Treatment (mg active ingredient/cm 2 ) Products, vendors, or commercial services mentioned or pictured in this seminar are for illustrative purposes only and are not meant Romero et al. -

WO 2011/094219 Al
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date , « i ... .. .. 4 August 2011 (04.08.2011) WO 2011/094219 Al (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every A01N 25/30 (2006.01) A01N 25/22 (2006.01) kind of national protection available): AE, AG, AL, AM, A 25/26 (2006.01) A01C 1/06 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, (21) International Application Number: DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, PCT/US201 1/022439 HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (22) International Filing Date: KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, 25 January 20 11 (25 .0 1.20 11) ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PE, PG, PH, PL, PT, RO, RS, RU, SC, SD, (25) Filing Language: English SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, (26) Publication Language: English TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 61/298,532 26 January 2010 (26.01 .2010) US kind of regional protection available): ARIPO (BW, GH, 61/321,966 8 April 2010 (08.04.2010) US GM, KE, LR, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, (72) Inventor; and TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, (71) Applicant : KROON, Hendrick-Andre [NL/US]; 24 EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, Sylvan Road North, Westport, Connecticut 06880 (US). -

China Releases New Maximum Residue Limits for Pesticides In
GB 2763-2016 THIS REPORT CONTAINS ASSESSMENTS OF COMMODITY AND TRADE ISSUES MADE BY USDA STAFF AND NOT NECESSARILY STATEMENTS OF OFFICIAL U.S. GOVERNMENT POLICY Voluntary - Public Date: 3/31/2017 GAIN Report Number: CH17016 China - Peoples Republic of Post: Beijing China Releases New Maximum Residue Limits for Pesticides in Food Report Categories: FAIRS Subject Report Approved By: Lisa Anderson Prepared By: FAS Staff Report Highlights: On December 18, 2016, the Chinese National Health and Family Planning Commission, Ministry of Agriculture, China Food and Drug Administration released the National Food Safety Standard - Maximum Residue Limits for Pesticides in Foods (GB 2763-2016). The standard will replace the current MRL Standard (GB 2763-2014) and will be implemented on June 18, 2017. This report provides an unofficial translation of the standard. Editors’ Note: The asterisk appearing in the MRL column means that the limit is a temporary MRL. A temporary MRL is usually set under the following four conditions: 1. The dietary risk assessment data is incomplete; 2. The Acceptable Daily Intake (ADI) is temporary (ADI is used as the basis for MRL setting); 3. There is no surveillance or analysis method for the MRL that complies with the standard requirements; 4. In emergency situations, the pesticide is approved to be used on un-registered crops. I GB 2763-2016 General Information: BEGIN TRANSLATION ICS 65.100 G 25 GB National Standard of the People’s Republic of China GB 2763—2016 Replacing GB 2763 - 2014 National food safety standard Maximum Residue Limits for Pesticides in Food General Information: National Health and Family Planning Commission Issued by: Ministry of Agriculture China Food and Drug Administration Issued on: 2016-12-18 Implementation:2017-06-18 II GB 2763-2016 Table of Content Preface ............................................................................................................................................................... -

(12) United States Patent (10) Patent No.: US 7.655,597 B1 Sanders (45) Date of Patent: Feb
USOO7655597 B1 (12) United States Patent (10) Patent No.: US 7.655,597 B1 Sanders (45) Date of Patent: Feb. 2, 2010 (54) PESTICIDE COMPOSITIONS INCLUDING 4,867,972 A 9, 1989 Girardeau et al. POLYMERC AIDUVANTS 6,093,679 A 7/2000 AZuma et al. 6,146,652 A 11/2000 Gore et al. 6,515,091 B2 2/2003 Sanders et al. (75) Inventor: John Larry Sanders, Leawood, KS 6,610,282 B1 8, 2003 Ghosh (US) 6,677,399 B2 1/2004 Herbert et al. 6,703,469 B2 3/2004 Sanders et al. (73) Assignee: Specialty Fertilizer Products, LLC, 6,706,666 B2 3/2004 HaSebe et al. Leawood, KS (US) 6,897,184 B2 5/2005 Kurita et al. 7,407,667 B2 8, 2008 Zerrer et al. (*) Notice: Subject to any disclaimer, the term of this 2004/0226331 A1 11/2004 Sanders et al. patent is extended or adjusted under 35 2004/0248741 A1 12/2004 Gotsche et al. U.S.C. 154(b) by 0 days. 2005/00904O2 A1* 4/2005 Dieing et al. ............... 504,361 2008.0167189 A1 7/2008 Oetter et al. (21) Appl. No.: 12/534,481 2008.0171658 A1 7/2008 Dyllick-Brenzinger et al. (22) Filed: Aug. 3, 2009 * cited by examiner Primary Examiner Johann R Richter (51) Int. Cl. Assistant Examiner—Andriae M Holt AOIN 25/00 (2006.01) (74) Attorney, Agent, or Firm Hovey Williams LLP AOIN 57/26 (2006.01) AOIN 25/28 (2006.01) (57) ABSTRACT (52) U.S. Cl. .................... 504/116.1; 504/206; 504/360; 504/361 Pesticidal compositions of improved effectiveness are pro (58) Field of Classification Search ................ -
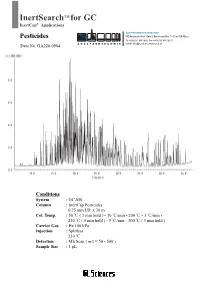
Inertsearch for GC
InertSearch TM for GC InertCap® Applications AUTHORIZED DISTRIBUTOR Pesticides MZ-Analysentechnik GmbH, Barcelona-Allee 17• D-55129 Mainz Tel +49 6131 880 96-0, Fax +49 6131 880 96-20 Data No. GA226-0904 e-mail: [email protected], www.mz-at. -

Suggested Insecticides for Managing Cotton Insects in the Lower Rio Grande Valley 2009
E-7A 7-09 Suggested Insecticides for Managing Cotton Insects in the Lower Rio Grande Valley 2009 This publication is to be used with E-7, Managing Cotton Insects in the Lower Rio Grande Valley—2009. Suggested Insecticides for Managing Cotton Insects in the Lower Rio Grande Valley D. L. Kerns and M. G Cattaneo* A committee of state and federal research scientists and Exten- these field tests met the minimum requirements as outlined in the sion specialists meets annually to review cotton pest management Guidelines for the Annual Entomology Research Review and Ex- research and management guidelines. Guidelines are revised at tension Guide Revision Conference. Products listed must conform this meeting to reflect the latest proven techniques for maximiz- to our performance standards and avoid undue environmental ing profits for the Texas cotton producer by optimizing inputs and consequences. production. Suggested insecticide use rates have exhibited sufficient effi- cacy in tests to be effective in providing adequate control in field Management of Cotton Pests situations. However, it is impossible to eliminate all risks. Condi- tions or circumstances that are unforeseen or unexpected may The proper management of cotton pests is dependent upon the result in less than satisfactory results. The Texas AgriLife Exten- use of pest management principles. Pest management does not sion Service will not assume responsibility for such risks. Such rely solely on insecticides. Therefore, the USER of this insert is responsibility shall be assumed by the user of this publication. strongly encouraged to refer to E-7 for discussion of pest biology, Suggested pesticides must be registered and labeled for use by the scouting techniques, economic thresholds, insecticide resistance Environmental Protection Agency and the Texas Department of management, conservation of existing natural control agents, Agriculture. -

Guideline Ver6.2 Se Ria L No. CAS RN® Substance Group 1 Substance
Guideline Ver6.2 Rank CAS RN® Substance Group 1 Substance Group 2 Substance Name 1=Prohibited No. Serial 2=Reduction 1 124-73-2 ozone depleting substances(Halons) Ethane, 1,2-dibromo-1,1,2,2-tetrafluoro- 1 2 25497-30-7 ozone depleting substances(Halons) Dibromotetrafluoroethane 1 3 27336-23-8 ozone depleting substances(Halons) Dibromotetrafluoroethane,compressed 1 4 353-59-3 ozone depleting substances(Halons) Methane, bromochlorodifluoro- 1 5 354-56-3 ozone depleting substances(Halons) Ethane, pentachlorofluoro- 1 6 75-61-6 ozone depleting substances(Halons) Methane, dibromodifluoro- 1 7 75-63-8 ozone depleting substances(Halons) Methane, bromotrifluoro- 1 8 110003-27-5 ozone depleting substances(HBFCs) 1,3-dibromo-1,1,2-trifluoropropane 1 9 111483-20-6 ozone depleting substances(HBFCs) 1-bromo-2,3-difluoropropane 1 10 124-72-1 ozone depleting substances(HBFCs) 2-bromo-1,1,1,2-tetrafluoroethane 1 11 148875-95-0 ozone depleting substances(HBFCs) C3H3FBr4 1 12 148875-98-3 ozone depleting substances(HBFCs) C3H2F2Br4 1 13 1511-62-2 ozone depleting substances(HBFCs) Methane, bromodifluoro- 1 14 172912-75-3 ozone depleting substances(HBFCs) HBFC-131 B3 Tribromofluoroethane 1 15 1800-81-3 ozone depleting substances(HBFCs) 1-bromo-1,2-difluoroethane 1 16 1868-53-7 ozone depleting substances(HBFCs) Dibromofluoromethane 1 17 1871-72-3 ozone depleting substances(HBFCs) FLUOROBROMOPROPANE 1 18 2252-78-0 ozone depleting substances(HBFCs) 1-Bromo-1,1,2,3,3,3-hexafluoropropane 1 19 2252-79-1 ozone depleting substances(HBFCs) 2-bromo-1,1,1,3,3,3-hexafluoropropane