X Chromosome Fact Sheet
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

X-Linked Recessive Inheritance
X-LINKED RECESSIVE Fact sheet 09 INHERITANCE This fact sheet talks about how genes affect our health when they follow a well understood pattern of genetic inheritance known as X-linked recessive inheritance. The exception to this rule applies to the genes carried on the sex chromosomes called X and Y. IN SUMMARY The genes in our DNA provide the instructions • Genes contain the instructions for for proteins, which are the building blocks of the growth and development. Some gene cells that make up our body. Although we all have variations or changes may mean that variation in our genes, sometimes this can affect the gene does not work properly or works in a different way that is harmful. how our bodies grow and develop. • A variation in a gene that causes a Generally, DNA variations that have no impact health or developmental condition on our health are called benign variants or is called a pathogenic variant or polymorphisms. These variants tend to be more mutation. common in people. Less commonly, variations can change the gene so that it sends a different • If a genetic condition happens when message. These changes may mean that the gene a gene on the X chromosome has does not work properly or works in a different way a variation, this is called X-linked that is harmful. A variation in a gene that causes inheritance. a health or developmental condition is called a • An X-linked recessive gene is a gene pathogenic variant or mutation. located on the X chromosome and affects males and females differently. -

Epigenetic Control of Mammalian Centromere Protein Binding: Does DNA Methylation Have a Role?
Journal of Cell Science 109, 2199-2206 (1996) 2199 Printed in Great Britain © The Company of Biologists Limited 1996 JCS3386 Epigenetic control of mammalian centromere protein binding: does DNA methylation have a role? Arthur R. Mitchell*, Peter Jeppesen, Linda Nicol†, Harris Morrison and David Kipling MRC Human Genetics Unit, Western General Hospital, Crewe Road, Edinburgh EH4 2XU, UK *Author for correspondence (internet [email protected]) †Present address: MRC Reproductive Biology Unit, Edinburgh, UK SUMMARY Chromosome 1 of the inbred mouse strain DBA/2 has a block of minor satellite DNA sequences on chromosome 1. polymorphism associated with the minor satellite DNA at The binding of the CENP-E protein does not appear to be its centromere. The more terminal block of satellite DNA affected by demethylation of the minor satellite sequences. sequences on this chromosome acts as the centromere as We present a model to explain these observations. This shown by the binding of CREST ACA serum, anti-CENP- model may also indicate the mechanism by which the B and anti-CENP-E polyclonal sera. Demethylation of the CENP-B protein recognises specific sites within the arrays minor satellite DNA sequences accomplished by growing of minor satellite DNA on mouse chromosomes. cells in the presence of the drug 5-aza-2′-deoxycytidine results in a redistribution of the CENP-B protein. This protein now binds to an enlarged area on the more terminal Key words: Centromere satellite DNA, Demethylation, Centromere block and in addition it now binds to the more internal antibody INTRODUCTION A common feature of many mammalian pericentromeric domains is that they contain families of repetitive DNA The centromere of mammalian chromosomes is recognised at sequences (Singer, 1982). -

X-Chromosome Meiotic Drive in Drosophila Simulans: a QTL Approach Reveals the Complex Polygenic Determinism of Paris Drive Suppression
Heredity (2019) 122:906–915 https://doi.org/10.1038/s41437-018-0163-1 ARTICLE X-chromosome meiotic drive in Drosophila simulans: a QTL approach reveals the complex polygenic determinism of Paris drive suppression 1 1,2 1 2 2 Cécile Courret ● Pierre R. Gérard ● David Ogereau ● Matthieu Falque ● Laurence Moreau ● Catherine Montchamp-Moreau1 Received: 31 July 2018 / Revised: 14 October 2018 / Accepted: 24 October 2018 / Published online: 5 December 2018 © The Genetics Society 2018 Abstract Meiotic drivers are selfish genetic elements that promote their own transmission into the gametes, which results in intragenomic conflicts. In the Paris sex-ratio system of Drosophila simulans, drivers located on the X chromosome prevent the segregation of the heterochromatic Y chromosome during meiosis II, and hence the production of Y-bearing sperm. The resulting sex-ratio bias strongly impacts population dynamics and evolution. Natural selection, which tends to restore an equal sex ratio, favors the emergence of resistant Y chromosomes and autosomal suppressors. This is the case in the Paris 1234567890();,: 1234567890();,: sex-ratio system where the drivers became cryptic in most of the natural populations of D. simulans. Here, we used a quantitative trait locus (QTL) mapping approach based on the analysis of 152 highly recombinant inbred lines (RILs) to investigate the genetic determinism of autosomal suppression. The RILs were derived from an advanced intercross between two parental lines, one showing complete autosomal suppression while the other one was sensitive to drive. The confrontation of RIL autosomes with a reference XSR chromosome allowed us to identify two QTLs on chromosome 2 and three on chromosome 3, with strong epistatic interactions. -

Sema4 Noninvasive Prenatal Select
Sema4 Noninvasive Prenatal Select Noninvasive prenatal testing with targeted genome counting 2 Autosomal trisomies 5 Trisomy 21 (Down syndrome) 6 Trisomy 18 (Edwards syndrome) 7 Trisomy 13 (Patau syndrome) 8 Trisomy 16 9 Trisomy 22 9 Trisomy 15 10 Sex chromosome aneuploidies 12 Monosomy X (Turner syndrome) 13 XXX (Trisomy X) 14 XXY (Klinefelter syndrome) 14 XYY 15 Microdeletions 17 22q11.2 deletion 18 1p36 deletion 20 4p16 deletion (Wolf-Hirschhorn syndrome) 20 5p15 deletion (Cri-du-chat syndrome) 22 15q11.2-q13 deletion (Angelman syndrome) 22 15q11.2-q13 deletion (Prader-Willi syndrome) 24 11q23 deletion (Jacobsen Syndrome) 25 8q24 deletion (Langer-Giedion syndrome) 26 Turnaround time 27 Specimen and shipping requirements 27 2 Noninvasive prenatal testing with targeted genome counting Sema4’s Noninvasive Prenatal Testing (NIPT)- Targeted Genome Counting analyzes genetic information of cell-free DNA (cfDNA) through a simple maternal blood draw to determine the risk for common aneuploidies, sex chromosomal abnormalities, and microdeletions, in addition to fetal gender, as early as nine weeks gestation. The test uses paired-end next-generation sequencing technology to provide higher depth across targeted regions. It also uses a laboratory-specific statistical model to help reduce false positive and false negative rates. The test can be offered to all women with singleton, twins and triplet pregnancies, including egg donor. The conditions offered are shown in below tables. For multiple gestation pregnancies, screening of three conditions -

Evolution on the X Chromosome: Unusual Patterns and Processes
REVIEWS Evolution on the X chromosome: unusual patterns and processes Beatriz Vicoso and Brian Charlesworth Abstract | Although the X chromosome is usually similar to the autosomes in size and cytogenetic appearance, theoretical models predict that its hemizygosity in males may cause unusual patterns of evolution. The sequencing of several genomes has indeed revealed differences between the X chromosome and the autosomes in the rates of gene divergence, patterns of gene expression and rates of gene movement between chromosomes. A better understanding of these patterns should provide valuable information on the evolution of genes located on the X chromosome. It could also suggest solutions to more general problems in molecular evolution, such as detecting selection and estimating mutational effects on fitness. Haldane’s rule Sex-chromosome systems have evolved independently the predictions of theoretical models of X-chromosome The disproportionate loss of many times, and have attracted much attention from evolution will shed light on the assumptions on which fitness to the heterogametic evolutionary geneticists. This work has mainly focused the models are based, such as the degree of dominance of sex in F1 hybrids between on the steps leading to the initial evolution of sex chro- mutations and the existence of opposing forces species. mosomes, and the genetic degeneration of Y and W of selection on males and females, leading to a better 1 Clade chromosomes . Here, we discuss the evolution of the understanding of the forces that shape the evolution of A group of species which share X chromosome in long-established sex-chromosome eukaryotic genomes. a common ancestor. -

X Chromosome-Linked and Mitochondrial Gene Control of Leber
Proc. Nati. Acad. Sci. USA Vol. 88, pp. 8198-8202, September 1991 Genetics X chromosome-linked and mitochondrial gene control of Leber hereditary optic neuropathy: Evidence from segregation analysis for dependence on X chromosome inactivation (two-locus inheritance/cytoplasmic inheritance/reduced penetrance) XIANGDONG BU AND JEROME I. ROTTER* Medical Genetics Birth Defects Center, Departments of Medicine and Pediatrics, Cedars-Sinai Medical Center and University of California School of Medicine, Los Angeles, CA 90048 Communicated by Giuseppe Attardi, July 1, 1991 ABSTRACT Leber hereditary optic neuropathy (LHON) linkage analysis, Chen et al. (5) excluded an X-linked gene has been shown to involve mutation(s) of mitochondrial DNA, alone as the cause for LHON. Earlier, Imai and Moriwaki (6) yet there remain several confusing aspects of Its inheritance not advanced the theory of cytoplasmic inheritance. The identi- explained by mitochondrial inheritance alone, including male fication of a mtDNA point mutation for LHON by Wallace et predominance, reduced penetrance, and a later age of onset in al. (3) and Singh et al. (4) subsequently proved the decades- females. By extending segregation analysis methods to disor- old hypothesis regarding the cytoplasmic inheritance of ders that involve both a mitochondrial and a nuclear gene LHON (6). As mentioned above, however, a mitochondrial locus, we show that the available pedigree data for LHON are mutation alone still cannot explain many ofthe features ofthe most consistent with a two-locus disorder, with one responsible transmission pattern of LHON, including the strong male gene being mitochondrial and the other nuclear and X chro- bias and the reduced penetrance of LHON in the maternal mosome-linked. -

Telomere-To-Telomere Assembly of a Complete Human X Chromosome W
https://doi.org/10.1038/s41586-020-2547-7 Accelerated Article Preview Telomere-to-telomere assembly of a W complete human X chromosome E VI Received: 30 July 2019 Karen H. Miga, Sergey Koren, Arang Rhie, Mitchell R. Vollger, Ariel Gershman, Andrey Bzikadze, Shelise Brooks, Edmund Howe, David Porubsky, GlennisE A. Logsdon, Accepted: 29 May 2020 Valerie A. Schneider, Tamara Potapova, Jonathan Wood, William Chow, Joel Armstrong, Accelerated Article Preview Published Jeanne Fredrickson, Evgenia Pak, Kristof Tigyi, Milinn Kremitzki,R Christopher Markovic, online 14 July 2020 Valerie Maduro, Amalia Dutra, Gerard G. Bouffard, Alexander M. Chang, Nancy F. Hansen, Amy B. Wilfert, Françoise Thibaud-Nissen, Anthony D. Schmitt,P Jon-Matthew Belton, Cite this article as: Miga, K. H. et al. Siddarth Selvaraj, Megan Y. Dennis, Daniela C. Soto, Ruta Sahasrabudhe, Gulhan Kaya, Telomere-to-telomere assembly of a com- Josh Quick, Nicholas J. Loman, Nadine Holmes, Matthew Loose, Urvashi Surti, plete human X chromosome. Nature Rosa ana Risques, Tina A. Graves Lindsay, RobertE Fulton, Ira Hall, Benedict Paten, https://doi.org/10.1038/s41586-020-2547-7 Kerstin Howe, Winston Timp, Alice Young, James C. Mullikin, Pavel A. Pevzner, (2020). Jennifer L. Gerton, Beth A. Sullivan, EvanL E. Eichler & Adam M. Phillippy C This is a PDF fle of a peer-reviewedI paper that has been accepted for publication. Although unedited, the Tcontent has been subjected to preliminary formatting. Nature is providing this early version of the typeset paper as a service to our authors and readers. The text andR fgures will undergo copyediting and a proof review before the paper is published in its fnal form. -

Structural and Numerical Changes of Chromosome X in Patients with Esophageal Atresia
European Journal of Human Genetics (2014) 22, 1077–1084 & 2014 Macmillan Publishers Limited All rights reserved 1018-4813/14 www.nature.com/ejhg ARTICLE Structural and numerical changes of chromosome X in patients with esophageal atresia Erwin Brosens*,1,2,5, Elisabeth M de Jong1,2,5, Tahsin Stefan Barakat3, Bert H Eussen1, Barbara D’haene4, Elfride De Baere4, Hannah Verdin4, Pino J Poddighe1, Robert-Jan Galjaard1, Joost Gribnau3, Alice S Brooks1, Dick Tibboel2 and Annelies de Klein1 Esophageal atresia with or without tracheoesophageal fistula (EA/TEF) is a relatively common birth defect often associated with additional congenital anomalies such as vertebral, anal, cardiovascular, renal and limb defects, the so-called VACTERL association. Yet, little is known about the causal genetic factors. Rare case reports of gastrointestinal anomalies in children with triple X syndrome prompted us to survey the incidence of structural and numerical changes of chromosome X in patients with EA/TEF. All available (n ¼ 269) karyotypes of our large (321) EA/TEF patient cohort were evaluated for X-chromosome anomalies. If sufficient DNA material was available, we determined genome-wide copy number profiles with SNP array and identified subtelomeric aberrations on the difficult to profile PAR1 region using telomere-multiplex ligation-dependent probe amplification. In addition, we investigated X-chromosome inactivation (XCI) patterns and mode of inheritance of detected aberrations in selected patients. Three EA/TEF patients had an additional maternally inherited X chromosome. These three female patients had normal random XCI patterns. Two male EA/TEF patients had small inherited duplications of the XY-linked SHOX (Short stature HOmeoboX-containing) locus. -
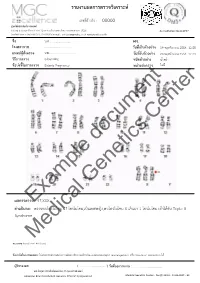
Example Document Medical Genetics Center
รายงานผลการตรวจววเคราะหย เลขทธรออางอนง : 00000 ศผนยยพวนธธศาสตรยการแพทยย 6/6 หมนม 6 ถนนสสขาภนบาล 5 ซอย 32 แขวงออเงนน เขตสายไหม กรสงเทพมหานคร 10220 Accreditation No.4148/57 โทรศกพทย/โทรสาร 086-8861203, 02-7920726 e-mail : [email protected] www.genetics.co.th ชชชอ น.ส............................. HN. ............. โรงพยาบาล .................................... ววนททชเกกบตววอยยาง 19 พฤศจนกายน 2558 12:55 แพทยยผผผสวชงตรวจ นพ.................................... ววนททชรวบตววอยยาง 20 พฤศจนกายน 2558 12:43 ววธทการตรวจ G-banding ชนวดตววอยยาง นนนาครรนา ขผอบยงชทชในการตรวจ Elderly Pregnancy หนยวยสยงตรวจ ไมมมธ Center document Genetics ผลการตรวจExample: 47,XXX คคาอธวบาย : ตรวจพบโครโมโซม 47 โครโมโซม,เปปนเพศหญนง,พบโครโมโซม X เกนนมา 1 โครโมโซม เขอาไดอกกบ Triple X Syndrome Medical หมายเหตธ: Band level 450 band ขผอจคากวดในการทดสอบ: ไมมสามารถตรวจสอบความผนดปกตนขนาดเลลกชนนด submicroscopic rearrangement หรรอ low-level mosaicism ไดอ ผผผรวบรองผล ( .................................. ) ววนททชออกรายงาน : ....................................... นพ.วทรยธทธ ประพวนธยพจนย,พบ.,วว.กธมารเวชศาสตรย American Board of Medical Genetics (Clinical Cytogenetics) Medical Genetics Center : Fo-QP-19/01 : 16-03-2015 : 02 เรียน อ.นพ...................................... ที่นับถือ ตามที่ผลตรวจโครโมโซมจากน ้าคร่้าของ นส........................ (HN .............., Lab no. ..............) พบ โครโมโซม X เกินมา 1 โครโมโซมนั น ความผิดปกติดังกล่าว เข้าได้กับกลุ่มอาการ trisomy X หรือ triple X syndrome ซึ่งเป็นความผิดปกติของโครโมโซมเพศที่พบได้บ่อยในทารกเพศหญิง โดยความผิดปกติของ โครโมโซมแบบนี -
Inside This Issue
Winter 2014 No. 77 Inside this issue Group News | Fundraising | Members’ Letters | One Family Living with Two Different Chromosome Disorders | Bristol Conference 2014 | Unique Leaflets | Christmas Card Order Form Sophie, Unique’s Chair of Trustees Dear Members, In the past month a few things have reminded me of why it is so important to make connections through Unique but also to draw support from other parents around us. I’ve just returned from Unique’s most recent family conference in Bristol where 150 of us parents and carers had a lovely time in workshops, meals and activities, chatting and watching our children milling around together like one big family since – although we had never met before – we have shared so many experiences in common. However in contrast I have also just met a new mum who has just moved to my area from far away with two toddlers, one with a rare joys of the internet, it is becoming easier to meet others with similar, chromosome disorder, who is starting from scratch with no even very rare, chromosome disorders around the world and to find professional, medical or social support. She reminds me of how yourself talking to them in the middle of the night about some lonely I felt when Max was newly diagnosed, when I knew no one interesting things our children share in common (obsession with with a disabled child let alone anyone with a rare chromosome catalogues, anyone?) And of course we also have an enormous disorder. Elsewhere our latest Unique Facebook group, Unique amount in common with so many parents of children with other Russia, is also just starting up – so far it includes just a small special needs or disabilities around us in our own communities who number of members sharing very different experiences to mine here will often be walking the same path as us. -

Prenatal Diagnosis by FISH of a 22Ql 1 Deletion in Two Families J Med Genet: First Published As 10.1136/Jmg.35.2.165 on 1 February 1998
I Med Genet 1998;35:165-168 165 Prenatal diagnosis by FISH of a 22ql 1 deletion in two families J Med Genet: first published as 10.1136/jmg.35.2.165 on 1 February 1998. Downloaded from Marie-France Portnoi, Nicole Joye, Marie Gonzales, Suzanne Demczuk, Laurent Fermont, Gilles Gaillard, Guy Bercau, Genevieve Morlier, Jean-Louis Taillemite Abstract balanced translocation t(I 1;22)(q23;ql 1) was We report on prenatal diagnosis by FISH shown in the father's karyotype; in the second of a sporadic 22qll deletion associated case trisomy X was associated with a 22ql 1 with DiGeorge syndrome (DGS) in two deletion in the fetus. fetuses after an obstetric ultrasonographic examination detected cardiac anomalies, Materials and methods an interrupted aortic arch in case 1 and KARYOTYPING AND FISH tetralogy of Fallot in case 2. The parents Fetal blood samples were obtained for karyo- decided to terminate the pregnancies. At type analysis and FISH. Cells were harvested necropsy, fetal examination showed char- from cultures of phytohaemagglutinin stimu- acteristic facial dysmorphism associated lated lymphocytes and spread onto slides for with congenital malformations, confirm- the production of G banded or R banded chro- ing full DGS in both fetuses. In addition to mosomes. the 22ql 1 deletion, trisomy X was found in FISH of metaphase chromosomes using dig- the second fetus and a reciprocal balanced oxigenin labelled cosmid probes D22S75 translocation t(l 1 ;22) (q23;ql 1) was found (N25, ONCOR) from the DGS chromosome in the clinically normal father of case 1. region was carried out basically according to These findings highlight the importance Pinkel et al.' This probe was premixed with a of performing traditional cytogenetic control cosmid (D22S39) in 22q13.3 facilitat- analysis and FISH in pregnancies with a ing chromosome identification. -

Double Aneuploidy in Down Syndrome
Chapter 6 Double Aneuploidy in Down Syndrome Fatma Soylemez Additional information is available at the end of the chapter http://dx.doi.org/10.5772/60438 Abstract Aneuploidy is the second most important category of chromosome mutations relat‐ ing to abnormal chromosome number. It generally arises by nondisjunction at ei‐ ther the first or second meiotic division. However, the existence of two chromosomal abnormalities involving both autosomal and sex chromosomes in the same individual is relatively a rare phenomenon. The underlying mechanism in‐ volved in the formation of double aneuploidy is not well understood. Parental ori‐ gin is studied only in a small number of cases and both nondisjunctions occurring in a single parent is an extremely rare event. This chapter reviews the characteristics of double aneuploidies in Down syndrome have been discussed in the light of the published reports. Keywords: Double aneuploidy, Down Syndrome, Klinefelter Syndrome, Chromo‐ some abnormalities 1. Introduction With the discovery in 1956 that the correct chromosome number in humans is 46, the new area of clinical cytogenetic began its rapid growth. Several major chromosomal syndromes with altered numbers of chromosomes were reported, such as Down syndrome (trisomy 21), Turner syndrome (45,X) and Klinefelter syndrome (47,XXY). Since then it has been well established that chromosome abnormalities contribute significantly to genetic disease resulting in reproductive loss, infertility, stillbirths, congenital anomalies, abnormal sexual development, mental retardation and pathogenesis of malignancy [1]. Clinical features of patients with common autosomal or sex chromosome aneuploidy is shown in Table 1. © 2015 The Author(s). Licensee InTech. This chapter is distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.