Structural and Numerical Changes of Chromosome X in Patients with Esophageal Atresia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Title of the Article
Mechanism-Anchored Profiling Derived from Epigenetic Networks Predicts Outcome in Acute Lymphoblastic Leukemia Xinan Yang, PhD1, Yong Huang, MD1, James L Chen, MD1, Jianming Xie, MSc2, Xiao Sun, PhD2, Yves A Lussier, MD1,3,4§ 1Center for Biomedical Informatics and Section of Genetic Medicine, Department of Medicine, The University of Chicago, Chicago, IL 60637 USA 2State Key Laboratory of Bioelectronics, Southeast University, 210096 Nanjing, P.R.China 3The University of Chicago Cancer Research Center, and The Ludwig Center for Metastasis Research, The University of Chicago, Chicago, IL 60637 USA 4The Institute for Genomics and Systems Biology, and the Computational Institute, The University of Chicago, Chicago, IL 60637 USA §Corresponding author Email addresses: XY: [email protected] YH: [email protected] JC: [email protected] JX: [email protected] XS: [email protected] YL: [email protected] - 1 - Abstract Background Current outcome predictors based on “molecular profiling” rely on gene lists selected without consideration for their molecular mechanisms. This study was designed to demonstrate that we could learn about genes related to a specific mechanism and further use this knowledge to predict outcome in patients – a paradigm shift towards accurate “mechanism-anchored profiling”. We propose a novel algorithm, PGnet, which predicts a tripartite mechanism-anchored network associated to epigenetic regulation consisting of phenotypes, genes and mechanisms. Genes termed as GEMs in this network meet all of the following criteria: (i) they are co-expressed with genes known to be involved in the biological mechanism of interest, (ii) they are also differentially expressed between distinct phenotypes relevant to the study, and (iii) as a biomodule, genes correlate with both the mechanism and the phenotype. -

Instability of the Pseudoautosomal Boundary in House Mice
Genetics: Early Online, published on April 26, 2019 as 10.1534/genetics.119.302232 Article/Investigation Instability of the pseudoautosomal boundary in house mice Andrew P Morgan∗, Timothy A Bell∗, James J Crowley∗; y; z, and Fernando Pardo-Manuel de Villena∗ ∗Department of Genetics and Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC yDepartment of Psychiatry, University of North Carolina, Chapel Hill, NC zDepartment of Clinical Neuroscience, Karolinska Institutet, Stockholm, Sweden April 26, 2019 Corresponding authors: Andrew P Morgan ([email protected]) Fernando Pardo-Manuel de Villena ([email protected]) 5049C Genetic Medicine Building Department of Genetics University of North Carolina Chapel Hill NC 27599-7264 Keywords: meiosis, recombination, pseudoautosomal region, sex chromosome evolution 1 Copyright 2019. Abstract Faithful segregation of homologous chromosomes at meiosis requires pairing and recombination. In taxa with dimorphic sex chromosomes, pairing between them in the heterogametic sex is limited to a narrow inter- val of residual sequence homology known as the pseudoautosomal region (PAR). Failure to form the obligate crossover in the PAR is associated with male infertility in house mice (Mus musculus) and humans. Yet despite this apparent functional constraint, the boundary and organization of the PAR is highly variable in mammals, and even between subspecies of mice. Here we estimate the genetic map in a previously-documented ex- pansion of the PAR in the Mus musculus castaneus subspecies and show that the local recombination rate is 100-fold higher than the autosomal background. We identify an independent shift in the PAR boundary in the Mus musculus musculus subspecies and show that it involves a complex rearrangement but still recombines in heterozygous males. -

Sema4 Noninvasive Prenatal Select
Sema4 Noninvasive Prenatal Select Noninvasive prenatal testing with targeted genome counting 2 Autosomal trisomies 5 Trisomy 21 (Down syndrome) 6 Trisomy 18 (Edwards syndrome) 7 Trisomy 13 (Patau syndrome) 8 Trisomy 16 9 Trisomy 22 9 Trisomy 15 10 Sex chromosome aneuploidies 12 Monosomy X (Turner syndrome) 13 XXX (Trisomy X) 14 XXY (Klinefelter syndrome) 14 XYY 15 Microdeletions 17 22q11.2 deletion 18 1p36 deletion 20 4p16 deletion (Wolf-Hirschhorn syndrome) 20 5p15 deletion (Cri-du-chat syndrome) 22 15q11.2-q13 deletion (Angelman syndrome) 22 15q11.2-q13 deletion (Prader-Willi syndrome) 24 11q23 deletion (Jacobsen Syndrome) 25 8q24 deletion (Langer-Giedion syndrome) 26 Turnaround time 27 Specimen and shipping requirements 27 2 Noninvasive prenatal testing with targeted genome counting Sema4’s Noninvasive Prenatal Testing (NIPT)- Targeted Genome Counting analyzes genetic information of cell-free DNA (cfDNA) through a simple maternal blood draw to determine the risk for common aneuploidies, sex chromosomal abnormalities, and microdeletions, in addition to fetal gender, as early as nine weeks gestation. The test uses paired-end next-generation sequencing technology to provide higher depth across targeted regions. It also uses a laboratory-specific statistical model to help reduce false positive and false negative rates. The test can be offered to all women with singleton, twins and triplet pregnancies, including egg donor. The conditions offered are shown in below tables. For multiple gestation pregnancies, screening of three conditions -

Predicting Clinical Response to Treatment with a Soluble Tnf-Antagonist Or Tnf, Or a Tnf Receptor Agonist
(19) TZZ _ __T (11) EP 2 192 197 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 02.06.2010 Bulletin 2010/22 C12Q 1/68 (2006.01) (21) Application number: 08170119.5 (22) Date of filing: 27.11.2008 (84) Designated Contracting States: (72) Inventor: The designation of the inventor has not AT BE BG CH CY CZ DE DK EE ES FI FR GB GR yet been filed HR HU IE IS IT LI LT LU LV MC MT NL NO PL PT RO SE SI SK TR (74) Representative: Habets, Winand Designated Extension States: Life Science Patents AL BA MK RS PO Box 5096 6130 PB Sittard (NL) (71) Applicant: Vereniging voor Christelijk Hoger Onderwijs, Wetenschappelijk Onderzoek en Patiëntenzorg 1081 HV Amsterdam (NL) (54) Predicting clinical response to treatment with a soluble tnf-antagonist or tnf, or a tnf receptor agonist (57) The invention relates to methods for predicting a clinical response to a therapy with a soluble TNF antagonist, TNF or a TNF receptor agonist and a kit for use in said methods. EP 2 192 197 A1 Printed by Jouve, 75001 PARIS (FR) EP 2 192 197 A1 Description [0001] The invention relates to methods for predicting a clinical response to a treatment with a soluble TNF antagonist, with TNF or a TNF receptor agonist using expression levels of genes of the Type I INF pathway and a kit for use in said 5 methods. In another aspect, the invention relates to a method for evaluating a pharmacological effect of a treatment with a soluble TNF antagonist, TNF or a TNF receptor agonist. -

A Molecular Switch for Photoperiod Responsiveness in Mammals
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology 20, 2193–2198, December 21, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2010.10.048 Report A Molecular Switch for Photoperiod Responsiveness in Mammals Hugues Dardente,1,* Cathy A. Wyse,1 Mike J. Birnie,1 type 2 thyroid hormone deiodinase (Dio2) expression in the Sandrine M. Dupre´,2 Andrew S.I. Loudon,2 hypothalamus [6]. This enzyme is a gatekeeper for the effects Gerald A. Lincoln,3 and David G. Hazlerigg1,* of thyroid hormone in the hypothalamus, controlling the avail- 1Institute of Biological and Environmental Sciences, Zoology ability of the active form of thyroid hormone, triiodothyronine, Building, Tillydrone Avenue, University of Aberdeen, and dictating the expression of summer phenotypes [7, 8]. Aberdeen AB24 2TZ, UK A unique feature of mammalian seasonal biology [2–5] is that 2Faculty of Life Sciences, University of Manchester, these effects of day length on PT function depend on the pineal Manchester M13 9PT, UK hormone melatonin, which acts via a high density of melatonin 3Queen’s Medical Research Institute, University of Edinburgh, receptors localized in the PT [ 9, 10]. Melatonin is a circadian Edinburgh EH16 4SB, UK signal, with a nocturnal waveform proportional to the length of the night, and consequently, in the PT, melatonin controls the rhythmical expression of multiple transcription factors Summary implicated in circadian function [11–15]. Hence, we hypothe- sized that photoperiodic effects on TSHb expression in the Seasonal synchronization based on day length (photope- PT are initiated through melatonin’s effects on circadian tran- riod) allows organisms to anticipate environmental change. -
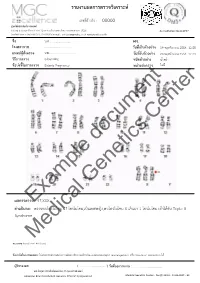
Example Document Medical Genetics Center
รายงานผลการตรวจววเคราะหย เลขทธรออางอนง : 00000 ศผนยยพวนธธศาสตรยการแพทยย 6/6 หมนม 6 ถนนสสขาภนบาล 5 ซอย 32 แขวงออเงนน เขตสายไหม กรสงเทพมหานคร 10220 Accreditation No.4148/57 โทรศกพทย/โทรสาร 086-8861203, 02-7920726 e-mail : [email protected] www.genetics.co.th ชชชอ น.ส............................. HN. ............. โรงพยาบาล .................................... ววนททชเกกบตววอยยาง 19 พฤศจนกายน 2558 12:55 แพทยยผผผสวชงตรวจ นพ.................................... ววนททชรวบตววอยยาง 20 พฤศจนกายน 2558 12:43 ววธทการตรวจ G-banding ชนวดตววอยยาง นนนาครรนา ขผอบยงชทชในการตรวจ Elderly Pregnancy หนยวยสยงตรวจ ไมมมธ Center document Genetics ผลการตรวจExample: 47,XXX คคาอธวบาย : ตรวจพบโครโมโซม 47 โครโมโซม,เปปนเพศหญนง,พบโครโมโซม X เกนนมา 1 โครโมโซม เขอาไดอกกบ Triple X Syndrome Medical หมายเหตธ: Band level 450 band ขผอจคากวดในการทดสอบ: ไมมสามารถตรวจสอบความผนดปกตนขนาดเลลกชนนด submicroscopic rearrangement หรรอ low-level mosaicism ไดอ ผผผรวบรองผล ( .................................. ) ววนททชออกรายงาน : ....................................... นพ.วทรยธทธ ประพวนธยพจนย,พบ.,วว.กธมารเวชศาสตรย American Board of Medical Genetics (Clinical Cytogenetics) Medical Genetics Center : Fo-QP-19/01 : 16-03-2015 : 02 เรียน อ.นพ...................................... ที่นับถือ ตามที่ผลตรวจโครโมโซมจากน ้าคร่้าของ นส........................ (HN .............., Lab no. ..............) พบ โครโมโซม X เกินมา 1 โครโมโซมนั น ความผิดปกติดังกล่าว เข้าได้กับกลุ่มอาการ trisomy X หรือ triple X syndrome ซึ่งเป็นความผิดปกติของโครโมโซมเพศที่พบได้บ่อยในทารกเพศหญิง โดยความผิดปกติของ โครโมโซมแบบนี -
Inside This Issue
Winter 2014 No. 77 Inside this issue Group News | Fundraising | Members’ Letters | One Family Living with Two Different Chromosome Disorders | Bristol Conference 2014 | Unique Leaflets | Christmas Card Order Form Sophie, Unique’s Chair of Trustees Dear Members, In the past month a few things have reminded me of why it is so important to make connections through Unique but also to draw support from other parents around us. I’ve just returned from Unique’s most recent family conference in Bristol where 150 of us parents and carers had a lovely time in workshops, meals and activities, chatting and watching our children milling around together like one big family since – although we had never met before – we have shared so many experiences in common. However in contrast I have also just met a new mum who has just moved to my area from far away with two toddlers, one with a rare joys of the internet, it is becoming easier to meet others with similar, chromosome disorder, who is starting from scratch with no even very rare, chromosome disorders around the world and to find professional, medical or social support. She reminds me of how yourself talking to them in the middle of the night about some lonely I felt when Max was newly diagnosed, when I knew no one interesting things our children share in common (obsession with with a disabled child let alone anyone with a rare chromosome catalogues, anyone?) And of course we also have an enormous disorder. Elsewhere our latest Unique Facebook group, Unique amount in common with so many parents of children with other Russia, is also just starting up – so far it includes just a small special needs or disabilities around us in our own communities who number of members sharing very different experiences to mine here will often be walking the same path as us. -

Transcription Mediated Insulation and Interference Direct Gene Cluster
1 Transcription mediated insulation and interference direct gene 2 cluster expression switches 3 Tania Nguyen1, Harry Fischl1#, Françoise S. Howe1#, Ronja Woloszczuk1#, Ana Serra 4 Barros1*, Zhenyu Xu2*, David Brown1, Struan C. Murray1, Simon Haenni1, James M. 5 Halstead1, Leigh O’Connor1, Gergana Shipkovenska1, Lars M. Steinmetz2 and Jane Mellor1§ 6 1Department of Biochemistry, South Parks Road, Oxford, OX1 3QU, UK 7 2European Molecular Biology Laboratory (EMBL), Genome Biology Unit, 69117 Heidelberg, 8 Germany 9 10 . 11 # , * Equal contribution 12 § Corresponding author: [email protected] 13 14 Competing interests statement: JM is an advisor to Oxford Biodynamics Ltd and Sibelius Ltd and sits 15 on the board of Chronos Therapeutics Ltd. OBD supported this work but have no commercial 16 interest in the study design, data collection and interpretation, or the decision to submit the work 17 for publication. 1 18 Abstract 19 In yeast, many tandemly arranged genes show peak expression in different phases of the 20 metabolic cycle (YMC) or in different carbon sources, indicative of regulation by a bi- 21 modal switch, but it is not clear how these switches are controlled. Using native 22 elongating transcript analysis (NET-seq), we show that transcription itself is a component 23 of bi-modal switches, facilitating reciprocal expression in gene clusters. HMS2, encoding a 24 growth-regulated transcription factor, switches between sense- or antisense-dominant 25 states that also coordinate up- and down-regulation of transcription at neighbouring 26 genes. Engineering HMS2 reveals alternative mono-, di- or tri-cistronic and antisense 27 transcription units (TUs), using different promoter and terminator combinations, that 28 underlie state-switching. -

273.Full-Text.Pdf
Copyright 2001 by the Genetics Society of America The te¯on Gene Is Required for Maintenance of Autosomal Homolog Pairing at Meiosis I in Male Drosophila melanogaster John E. Tomkiel,* Barbara T. Wakimoto² and Albert Briscoe, Jr.* *Center for Molecular Medicine and Genetics, Wayne State University, Detroit, Michigan 48202 and ²Departments of Zoology and Genetics, University of Washington, Seattle, Washington 98195 Manuscript received July 10, 2000 Accepted for publication September 29, 2000 ABSTRACT In recombination-pro®cient organisms, chiasmata appear to mediate associations between homologs at metaphase of meiosis I. It is less clear how homolog associations are maintained in organisms that lack recombination, such as male Drosophila. In lieu of chiasmata and synaptonemal complexes, there must be molecules that balance poleward forces exerted across homologous centromeres. Here we describe the genetic and cytological characterization of four EMS-induced mutations in te¯on (tef), a gene involved in this process in Drosophila melanogaster. All four alleles are male speci®c and cause meiosis I-speci®c nondisjunction of the autosomes. They do not measurably perturb sex chromosome segregation, suggesting that there are differences in the genetic control of autosome and sex chromosome segregation in males. Meiotic transmission of univalent chromosomes is unaffected in tef mutants, implicating the tef product in a pairing-dependent process. The segregation of translocations between sex chromosomes and autosomes is altered in tef mutants in a manner that supports this hypothesis. Consistent with these genetic observations, cytological examination of meiotic chromosomes suggests a role of tef in regulating or mediating pairing of autosomal bivalents at meiosis I. -

Prenatal Diagnosis by FISH of a 22Ql 1 Deletion in Two Families J Med Genet: First Published As 10.1136/Jmg.35.2.165 on 1 February 1998
I Med Genet 1998;35:165-168 165 Prenatal diagnosis by FISH of a 22ql 1 deletion in two families J Med Genet: first published as 10.1136/jmg.35.2.165 on 1 February 1998. Downloaded from Marie-France Portnoi, Nicole Joye, Marie Gonzales, Suzanne Demczuk, Laurent Fermont, Gilles Gaillard, Guy Bercau, Genevieve Morlier, Jean-Louis Taillemite Abstract balanced translocation t(I 1;22)(q23;ql 1) was We report on prenatal diagnosis by FISH shown in the father's karyotype; in the second of a sporadic 22qll deletion associated case trisomy X was associated with a 22ql 1 with DiGeorge syndrome (DGS) in two deletion in the fetus. fetuses after an obstetric ultrasonographic examination detected cardiac anomalies, Materials and methods an interrupted aortic arch in case 1 and KARYOTYPING AND FISH tetralogy of Fallot in case 2. The parents Fetal blood samples were obtained for karyo- decided to terminate the pregnancies. At type analysis and FISH. Cells were harvested necropsy, fetal examination showed char- from cultures of phytohaemagglutinin stimu- acteristic facial dysmorphism associated lated lymphocytes and spread onto slides for with congenital malformations, confirm- the production of G banded or R banded chro- ing full DGS in both fetuses. In addition to mosomes. the 22ql 1 deletion, trisomy X was found in FISH of metaphase chromosomes using dig- the second fetus and a reciprocal balanced oxigenin labelled cosmid probes D22S75 translocation t(l 1 ;22) (q23;ql 1) was found (N25, ONCOR) from the DGS chromosome in the clinically normal father of case 1. region was carried out basically according to These findings highlight the importance Pinkel et al.' This probe was premixed with a of performing traditional cytogenetic control cosmid (D22S39) in 22q13.3 facilitat- analysis and FISH in pregnancies with a ing chromosome identification. -

Double Aneuploidy in Down Syndrome
Chapter 6 Double Aneuploidy in Down Syndrome Fatma Soylemez Additional information is available at the end of the chapter http://dx.doi.org/10.5772/60438 Abstract Aneuploidy is the second most important category of chromosome mutations relat‐ ing to abnormal chromosome number. It generally arises by nondisjunction at ei‐ ther the first or second meiotic division. However, the existence of two chromosomal abnormalities involving both autosomal and sex chromosomes in the same individual is relatively a rare phenomenon. The underlying mechanism in‐ volved in the formation of double aneuploidy is not well understood. Parental ori‐ gin is studied only in a small number of cases and both nondisjunctions occurring in a single parent is an extremely rare event. This chapter reviews the characteristics of double aneuploidies in Down syndrome have been discussed in the light of the published reports. Keywords: Double aneuploidy, Down Syndrome, Klinefelter Syndrome, Chromo‐ some abnormalities 1. Introduction With the discovery in 1956 that the correct chromosome number in humans is 46, the new area of clinical cytogenetic began its rapid growth. Several major chromosomal syndromes with altered numbers of chromosomes were reported, such as Down syndrome (trisomy 21), Turner syndrome (45,X) and Klinefelter syndrome (47,XXY). Since then it has been well established that chromosome abnormalities contribute significantly to genetic disease resulting in reproductive loss, infertility, stillbirths, congenital anomalies, abnormal sexual development, mental retardation and pathogenesis of malignancy [1]. Clinical features of patients with common autosomal or sex chromosome aneuploidy is shown in Table 1. © 2015 The Author(s). Licensee InTech. This chapter is distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. -

Female Polysomy-X and Systemic Lupus Erythematosus
Seminars in Arthritis and Rheumatism 43 (2014) 508–512 Contents lists available at ScienceDirect Seminars in Arthritis and Rheumatism journal homepage: www.elsevier.com/locate/semarthrit Female polysomy-X and systemic lupus erythematosus Mordechai Slae, MDa,n, Merav Heshin-Bekenstein, MDb, Ari Simckes, MDb, Gali Heimer, MDb, Dan Engelhard, MDb, Eli M. Eisenstein, MDc a Department of Pediatrics, University of Alberta Hospital, Edmonton, Alberta, Canada b Department of Pediatrics, Hadassah-Hebrew University Hospital at Ein Kerem, Jerusalem, Israel c Department of Pediatrics, Hadassah-Hebrew University Hospital at Mount-Scopus, Jerusalem, Israel article info abstract Objectives: Systemic lupus erythematosus (SLE) occurs more commonly in females than in males. Recent Keywords: evidence suggests that genetic factors transmitted by the X-chromosome may confer increased risk for Gene dosage autoimmune disease in general, and for SLE in particular. It is therefore possible that X-chromosome Lupus genetics polysomy might confer further increased risk for lupus. In addition to describing the clinical and Polysomy-X immunologic features of a young woman with polysomy-X and SLE, we sought to review all other Sex differences published cases associating female or male polysomy-X with SLE or other forms of autoimmunity. SLE Methods: We report a case of a prepubertal girl with polysomy-X and SLE. We performed a systemic X-chromosome literature review for cases of polysomy-X and SLE and summarize previously published cases. In addition, we reviewed reports concerning the possible association between SLE and other connective tissue diseases and male polysomy-X. Results: An 11-year-old girl with tetrasomy-X (48 XXXX karyotype) presented with prolonged fever.