Restoration of Keratinocytic Phenotypes in Autonomous Trisomy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

By Submitted in Partial Satisfaction of the Requirements for Degree of in In
Developments of Two Imaging based Technologies for Cell Biology Researches by Xiaowei Yan DISSERTATION Submitted in partial satisfaction of the requirements for degree of DOCTOR OF PHILOSOPHY in Biochemistry and Molecular Biology in the GRADUATE DIVISION of the UNIVERSITY OF CALIFORNIA, SAN FRANCISCO Approved: ______________________________________________________________________________Ronald Vale Chair ______________________________________________________________________________Jonathan Weissman ______________________________________________________________________________Orion Weiner ______________________________________________________________________________ ______________________________________________________________________________ Committee Members Copyright 2021 By Xiaowei Yan ii DEDICATION Everything happens for the best. To my family, who supported me with all their love. iii ACKNOWLEDGEMENTS The greatest joy of my PhD has been joining UCSF, working and learning with such a fantastic group of scientists. I am extremely grateful for all the support and mentorship I received and would like to thank: My mentor, Ron Vale, who is such a great and generous person. Thank you for showing me that science is so much fun and thank you for always giving me the freedom in pursuing my interest. I am grateful for all the guidance from you and thank you for always supporting me whenever I needed. You are a person full of wisdom, and I have been learning so much from you and your attitude to science, science community and even life will continue inspire me. Thank you for being my mentor and thank you for being such a great mentor. Everyone else in Vale lab, past and present, for making our lab a sweet home. I would like to give my special thank to Marvin (Marvin Tanenbaum) and Nico (Nico Stuurman), two other mentors for me in the lab. I would like to thank them for helping me adapt to our lab, for all the valuable advice and for all the happiness during the time that we work together. -

V45n4a03.Pdf
Montoya JC/et al/Colombia Médica - Vol. 45 Nº4 2014 (Oct-Dec) Colombia Médica colombiamedica.univalle.edu.co Original Article Global differential expression of genes located in the Down Syndrome Critical Region in normal human brain Expresión diferencial global de genes localizados en la Región Crítica del Síndrome de Down en el cerebro humano normal Julio Cesar Montoya1,3, Dianora Fajardo2, Angela Peña2 , Adalberto Sánchez1, Martha C Domínguez1,2, José María Satizábal1, Felipe García-Vallejo1,2 1 Department of Physiological Sciences, School of Basic Sciences, Faculty of Health, Universidad del Valle. 2 Laboratory of Molecular Biology and Pathogenesis LABIOMOL. Universidad del Valle, Cali, Colombia. 3 Faculty of Basic Sciences, Universidad Autónoma de Occidente, Cali, Colombia. Montoya JC , Fajardo D, Peña A , Sánchez A, Domínguez MC, Satizábal JM, García-Vallejo F.. Global differential expression of genes located in the Down Syndrome Critical Region in normal human brain. Colomb Med. 2014; 45(4): 154-61. © 2014 Universidad del Valle. This is an Open Access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Article history Abstract Resumen Background: The information of gene expression obtained from Introducción: La información de la expresión de genes consignada Received: 2 July 2014 Revised: 10 November 2014 databases, have made possible the extraction and analysis of data en bases de datos, ha permitido extraer y analizar información acerca Accepted: 19 December 2014 related with several molecular processes involving not only in procesos moleculares implicados tanto en la homeostasis cerebral y su brain homeostasis but its disruption in some neuropathologies; alteración en algunas neuropatologías. -

NICU Gene List Generator.Xlsx
Neonatal Crisis Sequencing Panel Gene List Genes: A2ML1 - B3GLCT A2ML1 ADAMTS9 ALG1 ARHGEF15 AAAS ADAMTSL2 ALG11 ARHGEF9 AARS1 ADAR ALG12 ARID1A AARS2 ADARB1 ALG13 ARID1B ABAT ADCY6 ALG14 ARID2 ABCA12 ADD3 ALG2 ARL13B ABCA3 ADGRG1 ALG3 ARL6 ABCA4 ADGRV1 ALG6 ARMC9 ABCB11 ADK ALG8 ARPC1B ABCB4 ADNP ALG9 ARSA ABCC6 ADPRS ALK ARSL ABCC8 ADSL ALMS1 ARX ABCC9 AEBP1 ALOX12B ASAH1 ABCD1 AFF3 ALOXE3 ASCC1 ABCD3 AFF4 ALPK3 ASH1L ABCD4 AFG3L2 ALPL ASL ABHD5 AGA ALS2 ASNS ACAD8 AGK ALX3 ASPA ACAD9 AGL ALX4 ASPM ACADM AGPS AMELX ASS1 ACADS AGRN AMER1 ASXL1 ACADSB AGT AMH ASXL3 ACADVL AGTPBP1 AMHR2 ATAD1 ACAN AGTR1 AMN ATL1 ACAT1 AGXT AMPD2 ATM ACE AHCY AMT ATP1A1 ACO2 AHDC1 ANK1 ATP1A2 ACOX1 AHI1 ANK2 ATP1A3 ACP5 AIFM1 ANKH ATP2A1 ACSF3 AIMP1 ANKLE2 ATP5F1A ACTA1 AIMP2 ANKRD11 ATP5F1D ACTA2 AIRE ANKRD26 ATP5F1E ACTB AKAP9 ANTXR2 ATP6V0A2 ACTC1 AKR1D1 AP1S2 ATP6V1B1 ACTG1 AKT2 AP2S1 ATP7A ACTG2 AKT3 AP3B1 ATP8A2 ACTL6B ALAS2 AP3B2 ATP8B1 ACTN1 ALB AP4B1 ATPAF2 ACTN2 ALDH18A1 AP4M1 ATR ACTN4 ALDH1A3 AP4S1 ATRX ACVR1 ALDH3A2 APC AUH ACVRL1 ALDH4A1 APTX AVPR2 ACY1 ALDH5A1 AR B3GALNT2 ADA ALDH6A1 ARFGEF2 B3GALT6 ADAMTS13 ALDH7A1 ARG1 B3GAT3 ADAMTS2 ALDOB ARHGAP31 B3GLCT Updated: 03/15/2021; v.3.6 1 Neonatal Crisis Sequencing Panel Gene List Genes: B4GALT1 - COL11A2 B4GALT1 C1QBP CD3G CHKB B4GALT7 C3 CD40LG CHMP1A B4GAT1 CA2 CD59 CHRNA1 B9D1 CA5A CD70 CHRNB1 B9D2 CACNA1A CD96 CHRND BAAT CACNA1C CDAN1 CHRNE BBIP1 CACNA1D CDC42 CHRNG BBS1 CACNA1E CDH1 CHST14 BBS10 CACNA1F CDH2 CHST3 BBS12 CACNA1G CDK10 CHUK BBS2 CACNA2D2 CDK13 CILK1 BBS4 CACNB2 CDK5RAP2 -

USP16 Counteracts Mono-Ubiquitination of Rps27a And
RESEARCH ARTICLE USP16 counteracts mono-ubiquitination of RPS27a and promotes maturation of the 40S ribosomal subunit Christian Montellese1†, Jasmin van den Heuvel1,2, Caroline Ashiono1, Kerstin Do¨ rner1,2, Andre´ Melnik3‡, Stefanie Jonas1§, Ivo Zemp1, Paola Picotti3, Ludovic C Gillet1, Ulrike Kutay1* 1Institute of Biochemistry, ETH Zurich, Zurich, Switzerland; 2Molecular Life Sciences Ph.D. Program, Zurich, Switzerland; 3Institute of Molecular Systems Biology, ETH Zurich, Zurich, Switzerland Abstract Establishment of translational competence represents a decisive cytoplasmic step in the biogenesis of 40S ribosomal subunits. This involves final 18S rRNA processing and release of residual biogenesis factors, including the protein kinase RIOK1. To identify novel proteins promoting the final maturation of human 40S subunits, we characterized pre-ribosomal subunits trapped on RIOK1 by mass spectrometry, and identified the deubiquitinase USP16 among the captured factors. We demonstrate that USP16 constitutes a component of late cytoplasmic pre-40S subunits that promotes the removal of ubiquitin from an internal lysine of ribosomal protein *For correspondence: RPS27a/eS31. USP16 deletion leads to late 40S subunit maturation defects, manifesting in [email protected] incomplete processing of 18S rRNA and retarded recycling of late-acting ribosome biogenesis Present address: †CSL Behring, factors, revealing an unexpected contribution of USP16 to the ultimate step of 40S synthesis. CSL Biologics Research Center, Finally, ubiquitination of RPS27a appears to depend on active translation, pointing at a potential ‡ Bern, Switzerland; MSD Merck connection between 40S maturation and protein synthesis. Sharp & Dohme AG, Lucerne, Switzerland; §Institute of Molecular Biology and Biophysics, ETH Zurich, Zurich, Switzerland Introduction Ribosomes stand at the center of translation in all kingdoms of life, catalyzing the synthesis of pro- Competing interests: The teins by reading a messenger RNA (mRNA) template. -

Role of Senataxin in RNA: DNA Hybrids Resolution at DNA Double
Role of senataxin in RNA : DNA hybrids resolution at DNA double strand breaks Sarah Cohen To cite this version: Sarah Cohen. Role of senataxin in RNA : DNA hybrids resolution at DNA double strand breaks. Cellular Biology. Université Paul Sabatier - Toulouse III, 2019. English. NNT : 2019TOU30125. tel-02930730 HAL Id: tel-02930730 https://tel.archives-ouvertes.fr/tel-02930730 Submitted on 4 Sep 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. ����� ���� ������������������ ������������������������������������ ��������� ���!������"������ �� "��#�$�% ��� � ���� %��"���������"� ��� ��� � � � &������ ������������ ������� ����'������"�� � (����� �"�� ���"�� �������"�&)�����"����*���� (�+ "" ��"��� ��������"�����!��� ������������������,�,�$�,����-��.�� ���.�,����+&����-��" �������������,����/�������������� �������������������� �,�0�%�$�� ��� ���������,����-������� � �������0���+ � ����� ������'������� ������'������������ %����1�� ����� ����������������� /��������/�,�� ��� ���������� ������������������� ������������������������������ �� ����������������� -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

UC Davis Dermatology Online Journal
UC Davis Dermatology Online Journal Title Penicillamine-associated cutis laxa and milia en plaque - case report and review of cutaneous changes associated with penicillamine Permalink https://escholarship.org/uc/item/47p4d8zv Journal Dermatology Online Journal, 22(5) Authors Vajdi, Tina Lee, Wiggin Wu Paravar, Taraneh Publication Date 2016 DOI 10.5070/D3225030951 License https://creativecommons.org/licenses/by-nc-nd/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Volume 22 Number 5 May 2016 Photo Vignette Penicillamine-associated cutis laxa and milia en plaque - case report and review of cutaneous changes associated with penicillamine Tina Vajdi1, Wiggin Wu Lee2, Taraneh Paravar2 Dermatology Online Journal 22 (5): 12 1University of California, San Diego School of Medicine 2Department of Dermatology, University of California, San Diego Correspondence: Taraneh Paravar, MD Assistant Clinical Professor Department of Dermatology University of California, San Diego 8899 University Center Lane, Suite 350 San Diego, California 92122, USA Tel. (858) 657-8322 E-mail: [email protected] Abstract Penicillamine-induced skin changes are rare and include: hypersensitivity reactions, autoimmune reactions, and cutaneous elastoses. We report a case of a 73-year-old man with cystinuria taking penicillamine for over 50 years who presented with penicillamine-induced cutis laxa and milia en plaque. A brief review of penicillamine induced skin changes, specifically cutis laxa and milia en plaque, is presented. Key Words: penicillamine, elastic tissue, cystinuria, cutis laxa, milia en plaque Introduction Penicillamine is a chelating agent commonly used to treat cystinuria and Wilson disease. Cystinuria is a genetic disorder in which patients lack the cysteine amino acid transporter. -
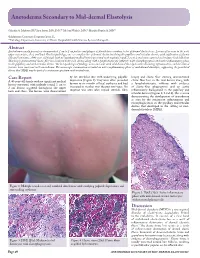
Anetoderma Secondary to Mid-Dermal Elastolysis
Anetoderma Secondary to Mid-dermal Elastolysis Gabriela A. Maloney, BS,* Jane James, MD, PhD,** Michael Welsch, MD,** Marylee Braniecki, MD** *Midwestern University, Downers Grove, IL **Pathology Department, University of Illinois Hospital & Health Sciences System, Chicago, IL Abstract Anetoderma usually presents as circumscribed, 1 cm to 2 cm patches and plaques of flaccid skin secondary to loss of dermal elastic tissue. Lesions often occur in the neck, upper extremities, chest, and back. On histopathology, one sees complete loss of dermal elastin involving the papillary and reticular dermis, with infiltration of plasma cells and histiocytes. A 40-year-old female with no significant medical history presented with multiple round, 1 cm to 2 cm lesions scattered on her upper back and chest. Skin biopsy demonstrated elastic-fiber loss localized to the mid-dermis along with a lymphohistiocytic infiltrate with elastophagocytosis and active inflammatory phase in the papillary and mid-reticular dermis. The histopathological findings were consistent with mid-dermal elastolysis with advancing inflammation, and the clinical features were consistent with anetoderma. The microscopic examination revealed an active inflammatory phase of mid-dermal elastolysis, supporting the postulated theory that MDE may be part of a continuous spectrum with anetoderma. Case Report by lax, wrinkled skin with underlying palpable biopsy and elastic-fiber staining demonstrated A 40-year-old female with no significant medical depression (Figure 1). They were often preceded elastic-fiber loss in the mid-dermis along with history presented with multiple round, 1 cm to by two to six months of local erythema and had a lymphohistiocytic infiltrate with evidence 2 cm lesions scattered throughout the upper increased in number over the past two years. -

Literature Mining Sustains and Enhances Knowledge Discovery from Omic Studies
LITERATURE MINING SUSTAINS AND ENHANCES KNOWLEDGE DISCOVERY FROM OMIC STUDIES by Rick Matthew Jordan B.S. Biology, University of Pittsburgh, 1996 M.S. Molecular Biology/Biotechnology, East Carolina University, 2001 M.S. Biomedical Informatics, University of Pittsburgh, 2005 Submitted to the Graduate Faculty of School of Medicine in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2016 UNIVERSITY OF PITTSBURGH SCHOOL OF MEDICINE This dissertation was presented by Rick Matthew Jordan It was defended on December 2, 2015 and approved by Shyam Visweswaran, M.D., Ph.D., Associate Professor Rebecca Jacobson, M.D., M.S., Professor Songjian Lu, Ph.D., Assistant Professor Dissertation Advisor: Vanathi Gopalakrishnan, Ph.D., Associate Professor ii Copyright © by Rick Matthew Jordan 2016 iii LITERATURE MINING SUSTAINS AND ENHANCES KNOWLEDGE DISCOVERY FROM OMIC STUDIES Rick Matthew Jordan, M.S. University of Pittsburgh, 2016 Genomic, proteomic and other experimentally generated data from studies of biological systems aiming to discover disease biomarkers are currently analyzed without sufficient supporting evidence from the literature due to complexities associated with automated processing. Extracting prior knowledge about markers associated with biological sample types and disease states from the literature is tedious, and little research has been performed to understand how to use this knowledge to inform the generation of classification models from ‘omic’ data. Using pathway analysis methods to better understand the underlying biology of complex diseases such as breast and lung cancers is state-of-the-art. However, the problem of how to combine literature- mining evidence with pathway analysis evidence is an open problem in biomedical informatics research. -

A Chromosome Level Genome of Astyanax Mexicanus Surface Fish for Comparing Population
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.06.189654; this version posted July 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Title 2 A chromosome level genome of Astyanax mexicanus surface fish for comparing population- 3 specific genetic differences contributing to trait evolution. 4 5 Authors 6 Wesley C. Warren1, Tyler E. Boggs2, Richard Borowsky3, Brian M. Carlson4, Estephany 7 Ferrufino5, Joshua B. Gross2, LaDeana Hillier6, Zhilian Hu7, Alex C. Keene8, Alexander Kenzior9, 8 Johanna E. Kowalko5, Chad Tomlinson10, Milinn Kremitzki10, Madeleine E. Lemieux11, Tina 9 Graves-Lindsay10, Suzanne E. McGaugh12, Jeff T. Miller12, Mathilda Mommersteeg7, Rachel L. 10 Moran12, Robert Peuß9, Edward Rice1, Misty R. Riddle13, Itzel Sifuentes-Romero5, Bethany A. 11 Stanhope5,8, Clifford J. Tabin13, Sunishka Thakur5, Yamamoto Yoshiyuki14, Nicolas Rohner9,15 12 13 Authors for correspondence: Wesley C. Warren ([email protected]), Nicolas Rohner 14 ([email protected]) 15 16 Affiliation 17 1Department of Animal Sciences, Department of Surgery, Institute for Data Science and 18 Informatics, University of Missouri, Bond Life Sciences Center, Columbia, MO 19 2 Department of Biological Sciences, University of Cincinnati, Cincinnati, OH 20 3 Department of Biology, New York University, New York, NY 21 4 Department of Biology, The College of Wooster, Wooster, OH 22 5 Harriet L. Wilkes Honors College, Florida Atlantic University, Jupiter FL 23 6 Department of Genome Sciences, University of Washington, Seattle, WA 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.07.06.189654; this version posted July 6, 2020. -

Dermatose Degenerativa Induzida Por D-Penicilamina Em Paciente Com Doença De Wilson
Revista SPDV 76(2) 2018; D-Penicillamine induced degenerative dermopathy; Rui Pedro Santos, Joana Gomes, Celeste Brito. Caso Clínico Dermatose Degenerativa Induzida por D-penicilamina em Paciente com Doença de Wilson Rui Pedro Santos1, Joana Gomes2, Celeste Brito2 1Interno de Dermatovenereologia/Resident, Dermatovenereology, Hospital de Braga, Braga, Portugal 2Especialista de Dermatovenereologia/Specialist of Dermatovenereology, Hospital de Braga, Braga, Portugal RESUMO – As dermatoses degenerativas induzidas por D-penicilamina incluem, entre outras, a elastose perfurante serpiginosa e o pseudo-pseudoxantoma elástico. A elastose perfurante serpiginosa é uma doença perfurante rara caracterizada pela elimi- nação transepidérmica de fibras elásticas anormais. Esta condição pode ser idiopática, reativa ou induzida por D-penicilamina, habitualmente utilizada para o tratamento da doença de Wilson, cistinúria, artrite reumatóide ou esclerose sistémica. Manifesta- ções cutâneas semelhantes a pseudoxantoma elástico mas sem história familiar e mutações do gene ABCC6 foram identificadas como sendo uma dermatose induzida por D-penicilamina e designada de pseudo-pseudoxantoma elástico. Descreve-se o caso de uma mulher de 17 anos tratada por vários anos com D-penicilamina para doença de Wilson, com pápulas assintomáticas, algumas cor de pele e hiperqueratósicas e outras macias e amareladas, na região cervical e face. A histopatolo- gia mostrou a eliminação transepidérmica de fibras elásticas espessadas, em forma de dentes de serra. Estes achados sugeriram uma dermopatia induzida por D-penicilamina e os autores consideraram o diagnóstico de elastose perfurante serpiginosa e pseudo-pseudoxantoma elástico no mesmo paciente. O fármaco foi alterado para acetato de zinco sem lesões novas, mas com manutenção das lesões existentes no seguimento a 1 ano. -

Aberrant Gene Expression: Diagnostic Markers and Therapeutic Targets for Pancreatic Cancer
ABERRANT GENE EXPRESSION: DIAGNOSTIC MARKERS AND THERAPEUTIC TARGETS FOR PANCREATIC CANCER Jeran Kent Stratford A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology. Chapel Hill 2014 Approved by: Jen Jen Yeh Channing J. Der Gary L. Johnson Adrienne D. Cox Timothy C. Elston © 2014 Jeran Kent Stratford ALL RIGHTS RESERVED ii ABSTRACT Jeran Kent Stratford: Aberrant gene expression: diagnostic markers and therapeutic targets for pancreatic cancer (Under the direction of Jen Jen Yeh and Channing J. Der) Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer and the fourth leading cause of cancer-related death in the United States. The overall median survival for patients diagnosed with PDAC is five to eight months. The poor outcome is due, in part, to a lack of disease-specific symptoms that can be used for early detection, and as such, most patients present with locally advanced or metastatic disease at the time of diagnosis. Therefore, the need for diagnostic tools is both great and urgent. Furthermore, current chemotherapies have low response rates and high toxicity, limiting their use, and there are currently no effective targeted therapies for PDAC. Therefore, a greater understanding of the underlying biology of pancreatic cancer is needed to identify tumor-specific vulnerabilities that can be therapeutically exploited. Pancreatic cancer development is driven by genomic changes that alter gene expression. Aberrant gene expression produces changes in protein expression, which in turn may confer growth advantages to the tumor; often the tumor then develops a dependency on continued aberrant gene and protein expression.