Package 'Splitstackshape'
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

At—At, Batch—Execute Commands at a Later Time
at—at, batch—execute commands at a later time at [–csm] [–f script] [–qqueue] time [date] [+ increment] at –l [ job...] at –r job... batch at and batch read commands from standard input to be executed at a later time. at allows you to specify when the commands should be executed, while jobs queued with batch will execute when system load level permits. Executes commands read from stdin or a file at some later time. Unless redirected, the output is mailed to the user. Example A.1 1 at 6:30am Dec 12 < program 2 at noon tomorrow < program 3 at 1945 pm August 9 < program 4 at now + 3 hours < program 5 at 8:30am Jan 4 < program 6 at -r 83883555320.a EXPLANATION 1. At 6:30 in the morning on December 12th, start the job. 2. At noon tomorrow start the job. 3. At 7:45 in the evening on August 9th, start the job. 4. In three hours start the job. 5. At 8:30 in the morning of January 4th, start the job. 6. Removes previously scheduled job 83883555320.a. awk—pattern scanning and processing language awk [ –fprogram–file ] [ –Fc ] [ prog ] [ parameters ] [ filename...] awk scans each input filename for lines that match any of a set of patterns specified in prog. Example A.2 1 awk '{print $1, $2}' file 2 awk '/John/{print $3, $4}' file 3 awk -F: '{print $3}' /etc/passwd 4 date | awk '{print $6}' EXPLANATION 1. Prints the first two fields of file where fields are separated by whitespace. 2. Prints fields 3 and 4 if the pattern John is found. -

Course Outline & Schedule
Course Outline & Schedule Call US 408-759-5074 or UK +44 20 7620 0033 Linux Advanced Shell Programming Tools Curriculum The Linux Foundation Course Code LASP Duration 3 Day Course Price $1,830 Course Description The Linux Advanced Shell Programming Tools course is designed to give delegates practical experience using a range of Linux tools to manipulate text and incorporate them into Linux shell scripts. The delegate will practise: Using the shell command line editor Backing up and restoring files Scheduling background jobs using cron and at Using regular expressions Editing text files with sed Using file comparison utilities Using the head and tail utilities Using the cut and paste utilities Using split and csplit Identifying and translating characters Sorting files Translating characters in a file Selecting text from files with the grep family of commands Creating programs with awk Course Modules Review of Shell Fundamentals (1 topic) ◾ Review of UNIX Commands Using Unix Shells (6 topics) ◾ Command line history and editing ◾ The Korn and POSIX shells Perpetual Solutions - Page 1 of 5 Course Outline & Schedule Call US 408-759-5074 or UK +44 20 7620 0033 ◾ The Bash shell ◾ Command aliasing ◾ The shell startup file ◾ Shell environment variables Redirection, Pipes and Filters (7 topics) ◾ Standard I/O and redirection ◾ Pipes ◾ Command separation ◾ Conditional execution ◾ Grouping Commands ◾ UNIX filters ◾ The tee command Backup and Restore Utilities (6 topics) ◾ Archive devices ◾ The cpio command ◾ The tar command ◾ The dd command ◾ Exercise: -

HEP Computing Part I Intro to UNIX/LINUX Adrian Bevan
HEP Computing Part I Intro to UNIX/LINUX Adrian Bevan Lectures 1,2,3 [email protected] 1 Lecture 1 • Files and directories. • Introduce a number of simple UNIX commands for manipulation of files and directories. • communicating with remote machines [email protected] 2 What is LINUX • LINUX is the operating system (OS) kernel. • Sitting on top of the LINUX OS are a lot of utilities that help you do stuff. • You get a ‘LINUX distribution’ installed on your desktop/laptop. This is a sloppy way of saying you get the OS bundled with lots of useful utilities/applications. • Use LINUX to mean anything from the OS to the distribution we are using. • UNIX is an operating system that is very similar to LINUX (same command names, sometimes slightly different functionalities of commands etc). – There are usually enough subtle differences between LINUX and UNIX versions to keep you on your toes (e.g. Solaris and LINUX) when running applications on multiple platforms …be mindful of this if you use other UNIX flavours. – Mac OS X is based on a UNIX distribution. [email protected] 3 Accessing a machine • You need a user account you should all have one by now • can then log in at the terminal (i.e. sit in front of a machine and type in your user name and password to log in to it). • you can also log in remotely to a machine somewhere else RAL SLAC CERN London FNAL in2p3 [email protected] 4 The command line • A user interfaces with Linux by typing commands into a shell. -

CESM2 Tutorial: Basic Modifications
CESM2 Tutorial: Basic Modifications Christine Shields August 15, 2017 CESM2 Tutorial: Basic Modifications: Review 1. We will use the CESM code located locally on Cheyenne, no need to checkout or download any input data. 2. We will run with resolution f19_g17: (atm/lnd = FV 1.9x2.5 ocn/ice=gx1v7) 3. Default scripts will automatically be configured for you using the code/script base prepared uniquely for this tutorial. 4. For On-site Tutorial ONLY: Please use compute nodes for compiling and login nodes for all other work, including submission. Please do NOT compile unless you have a compile card. To make the tutorial run smoothly for all, we need to regulate the model compiles. When you run from home, you don’t need to compile on the compute nodes. Tutorial Code and script base: /glade/p/cesm/tutorial/cesm2_0_alpha07c/ CESM2 Tutorial: Basic Modifications: Review 1. Log into Cheyenne 2. Execute create_newcase 3. Execute case.setup 4. Log onto compute node (compile_node.csH) 5. Compile model (case.build) 6. Exit compute node (type “exit”) 7. Run model (case.submit) This tutorial contains step by step instructions applicable to CESM2 (wHicH Has not been officially released yet). Documentation: Under construction! http://www.cesm.ucar.edu/models/cesm2.0/ Quick Start Guide: Under construction! http://cesm-development.github.io/cime/doc/build/html/index.Html For older releases, please see past tutorials. CESM2 Tutorial: Basic Modifications: Review: Creating a new case What is the Which Which model configuration ? Which set of components ? casename -

Xshell 6 User Guide Secure Terminal Emualtor
Xshell 6 User Guide Secure Terminal Emualtor NetSarang Computer, Inc. Copyright © 2018 NetSarang Computer, Inc. All rights reserved. Xshell Manual This software and various documents have been produced by NetSarang Computer, Inc. and are protected by the Copyright Act. Consent from the copyright holder must be obtained when duplicating, distributing or citing all or part of this software and related data. This software and manual are subject to change without prior notice for product functions improvement. Xlpd and Xftp are trademarks of NetSarang Computer, Inc. Xmanager and Xshell are registered trademarks of NetSarang Computer, Inc. Microsoft Windows is a registered trademark of Microsoft. UNIX is a registered trademark of AT&T Bell Laboratories. SSH is a registered trademark of SSH Communications Security. Secure Shell is a trademark of SSH Communications Security. This software includes software products developed through the OpenSSL Project and used in OpenSSL Toolkit. NetSarang Computer, Inc. 4701 Patrick Henry Dr. BLDG 22 Suite 137 Santa Clara, CA 95054 http://www.netsarang.com/ Contents About Xshell ............................................................................................................................................... 1 Key Functions ........................................................................................................... 1 Minimum System Requirements .................................................................................. 3 Install and Uninstall .................................................................................................. -

Other Useful Commands
Bioinformatics 101 – Lecture 2 Introduction to command line Alberto Riva ([email protected]), J. Lucas Boatwright ([email protected]) ICBR Bioinformatics Core Computing environments ▪ Standalone application – for local, interactive use; ▪ Command-line – local or remote, interactive use; ▪ Cluster oriented: remote, not interactive, highly parallelizable. Command-line basics ▪ Commands are typed at a prompt. The program that reads your commands and executes them is the shell. ▪ Interaction style originated in the 70s, with the first visual terminals (connections were slow…). ▪ A command consists of a program name followed by options and/or arguments. ▪ Syntax may be obscure and inconsistent (but efficient!). Command-line basics ▪ Example: to view the names of files in the current directory, use the “ls” command (short for “list”) ls plain list ls –l long format (size, permissions, etc) ls –l –t sort newest to oldest ls –l –t –r reverse sort (oldest to newest) ls –lrt options can be combined (in this case) ▪ Command names and options are case sensitive! File System ▪ Unix systems are centered on the file system. Huge tree of directories and subdirectories containing all files. ▪ Everything is a file. Unix provides a lot of commands to operate on files. ▪ File extensions are not necessary, and are not recognized by the system (but may still be useful). ▪ Please do not put spaces in filenames! Permissions ▪ Different privileges and permissions apply to different areas of the filesystem. ▪ Every file has an owner and a group. A user may belong to more than one group. ▪ Permissions specify read, write, and execute privileges for the owner, the group, everyone else. -

Conditioning Flyer
Winter Sports Condi�oning: Girls’ and Boys‘ Basketball, Wrestling, and Sideline Cheer can start condi�oning on October 19th. Exact dates and �mes will be sent out later. The first two weeks will be outdoors with no equipment. Fall Sports Condi�oning: Football, Golf, Cross Country, Volleyball, and Compe��on Cheer can start condi�oning on November 16th. Exact dates and �mes will be sent out later. The first two weeks will be outdoors with no equipment. Must Complete Before Condi�oning: • Must have a physical and the VHSL physical form completed on or a�er May 1st 2020 • Must have complete the Acknowledgement of the Concussion materials • Must have completed the Acknowledgement of Par�cipa�on in the Covid-19 parent mi�ga�on form h�ps://whs.iwcs.k12.va.us/ - this is the link to the WHS web page where the forms can be found. Winter Parent Mee�ng Dates and Zoom Links: • Sideline Cheer - October 12th at 6pm h�ps://us02web.zoom.us/j/89574054101?pwd=djJyMFZSeU9QeFpjdGd6VFB4NkZoQT09 • Wrestling - October 13th at 6pm h�ps://us02web.zoom.us/j/87806116997?pwd=c0RKa1I1NFU1T2ZZbWNUVVZqaG9rdz09 • Girls’ and Boys’ Basketball - October 14th at 6pm h�ps://us02web.zoom.us/j/86956809676?pwd=UWkycEJ2K0pBbW0zNk5tWmE0bkpuUT09 Fall Parent Mee�ng Dates and Zoom Links: • Football - November 9th at 6:00pm h�ps://us02web.zoom.us/j/81813330973?pwd=b0I3REJ0WUZtTUs4Z0o3RDNtNzd3dz09 • Cross Country and Golf - November 10th at 6:00pm h�ps://us02web.zoom.us/j/86072144126?pwd=U0FUb0M2a3dBaENIaDVRYmVBNW1KUT09 • Volleyball - November 12th at 6:00pm h�ps://us02web.zoom.us/j/82413556218?pwd=ZjdZdzZhODNVMHlVSk5kSk5CcjBwQT09 • Compe��on Cheer - November 12th at 7:30pm h�ps://us02web.zoom.us/j/81803664890?pwd=dWVDaHNZS0JTdXdWNlNrZkJoVi92UT09 The parent and student athlete must attend the zoom meeting or watch the recorded zoom session prior to attending conditioning. -
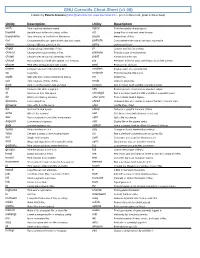
GNU Coreutils Cheat Sheet (V1.00) Created by Peteris Krumins ([email protected], -- Good Coders Code, Great Coders Reuse)
GNU Coreutils Cheat Sheet (v1.00) Created by Peteris Krumins ([email protected], www.catonmat.net -- good coders code, great coders reuse) Utility Description Utility Description arch Print machine hardware name nproc Print the number of processors base64 Base64 encode/decode strings or files od Dump files in octal and other formats basename Strip directory and suffix from file names paste Merge lines of files cat Concatenate files and print on the standard output pathchk Check whether file names are valid or portable chcon Change SELinux context of file pinky Lightweight finger chgrp Change group ownership of files pr Convert text files for printing chmod Change permission modes of files printenv Print all or part of environment chown Change user and group ownership of files printf Format and print data chroot Run command or shell with special root directory ptx Permuted index for GNU, with keywords in their context cksum Print CRC checksum and byte counts pwd Print current directory comm Compare two sorted files line by line readlink Display value of a symbolic link cp Copy files realpath Print the resolved file name csplit Split a file into context-determined pieces rm Delete files cut Remove parts of lines of files rmdir Remove directories date Print or set the system date and time runcon Run command with specified security context dd Convert a file while copying it seq Print sequence of numbers to standard output df Summarize free disk space setuidgid Run a command with the UID and GID of a specified user dir Briefly list directory -

Reference Guide Vmware Vcenter Server Heartbeat 5.5 Update 1
Administrator Guide VMware vCenter Server Heartbeat 6.3 Update 1 This document supports the version of each product listed and supports all subsequent versions until the document is replaced by a new edition. To check for more recent editions of this document, see http://www.vmware.com/support/pubs. EN-000562-01 Administrator Guide You can find the most up-to-date technical documentation on the VMware Web site at: http://www.vmware.com/support/ The VMware Web site also provides the latest product updates. If you have comments about this documentation, submit your feedback to: [email protected] Copyright © 2010 VMware, Inc. All rights reserved. This product is protected by U.S. and international copyright and intellectual property laws. VMware products are covered by one or more patents listed at http://www.vmware.com/go/patents. VMware is a registered trademark or trademark of VMware, Inc. in the United States and/or other jurisdictions. All other marks and names mentioned herein may be trademarks of their respective companies. VMware, Inc. 3401 Hillview Ave. Palo Alto, CA 94304 www.vmware.com 2 VMware, Inc. Contents About This Book 7 Getting Started 1 Introduction 11 Overview 11 vCenter Server Heartbeat Concepts 11 Architecture 11 Protection Levels 13 Communications 16 vCenter Server Heartbeat Switchover and Failover Processes 17 2 Configuring vCenter Server Heartbeat 21 Server Configuration Wizard 22 Configuring the Machine Identity 22 Configuring the Server Role 23 Configuring the Client Connection Port 23 Configuring Channel -

TEAL BOHO GLAM WEDDING CAKE with Jessica MV
TEAL BOHO GLAM WEDDING CAKE with Jessica MV Supplies and Resources THREE MACRAMES DESIGNS White modeling paste Crisco Cornstarch Extruder Single sided craft blade Cutting mat Fresh dry and damp towels Edible glue Paintbrush Global Sugar Art gold highlighter Faye Carhill signature gold lustre dust Lemon extract Baking paper (to put underneath the cake when you paint the patterns to avoid stains on the working counter) Non-slip pad (to keep the cake in place on the turntable) Turntable OMBRE CHEVRON IN BARGELLO ART Ruler Small pizza cutter Modeling paste tinted in 6 shades of teal Cutting mat White gum paste Edible glue Scale Paintbrush Crisco CK confectioner’s glaze Cornstarch Baking paper (to put underneath the cake when you glazethe patterns to Rolling pin avoid stains on the working counter) Kitchen Aid stand mixer with pasta roller & fettucine attachments Cake Jess., JSC cakejess.com 1 TEAL BOHO GLAM WEDDING CAKE with Jessica MV To tint the modeling paste, Jessica used: Supplies and Resources Wilton teal gel color Wilton moss green gel color Cutting mat CRACKLED TEXTURE AND RUFFLES 28-gauge white floral wire Dark teal, light teal, grey and white gum Green floral tape paste Squires Kitchen Multi-Flower Petal Crisco Cutter set 1, the 3 biggest cutters Cornstarch Big ball tool Extruder Small ball tool Rolling pin Giant rose petal veiners by Sugar Art Kitchen Aid stand mixer with pasta roller Studio attachment Edible glue Small pizza cutter Paintbrush Paintbrush Semi-sphere molds Edible glue Aluminum foil Global Sugar Art gold highlighter -

Reclaim Disk Space by Shrinking Files 49 from the Preceding Discussion It Should Be Clear That Cfsize and Csplit Are Two Very Different Tools
sysadmins from time to time are faced with the problem of reclaiming disk sandeep saHoRe space, a problem that lurks in the shadows waiting to buzz the pager into life. The typi- cal response is either to remove files or to compress them, or to invoke some combina- reclaim disk space tion of the two approaches. But there are many situations where the choice is not so by shrinking files cut-and-dried. Let’s say there is a file fill- Sandeep Sahore holds a Master’s degree in com- ing up a storage partition that cannot be puter science from the University of Toledo and removed because its data should be kept for has nearly 15 years of experience in the comput- ing industry. He specializes in low-level and C a rolling window of one year or because its programming, systems engineering, and system performance. contents are sensitive or because it is being [email protected] held open by a process. In such cases it is better to shrink the file in size instead of The source for cfsize.c may be found at http:// removing or compressing it. www.usenix.org/publications/login/2008-10/ cfsize.c. Having faced such scenarios a countless number of times I decided to write a program that would shrink files in size from the beginning or “head.” This program is called cfsize, which is short for “change file size”; it is described in detail in the sections that follow. cfsize is written in the C lan- guage for the UNIX platform, but it works equally well on Linux. -

Windows Tool Reference
AppendixChapter A1 Windows Tool Reference Windows Management Tools This appendix lists sets of Windows management, maintenance, configuration, and monitor- ing tools that you may not be familiar with. Some are not automatically installed by Windows Setup but instead are hidden away in obscure folders on your Windows Setup DVD or CD- ROM. Others must be downloaded or purchased from Microsoft. They can be a great help in using, updating, and managing Windows. We’ll discuss the following tool kits: ■ Standard Tools—Our pick of handy programs installed by Windows Setup that we think are unappreciated and not well-enough known. ■ Support Tools—A set of useful command-line and GUI programs that can be installed from your Windows Setup DVD or CD-ROM. ■ Value-Added Tools—Several more sets of utilities hidden away on the Windows Setup CD-ROM. ■ Windows Ultimate Extras and PowerToys for XP—Accessories that can be downloaded for free from microsoft.com. The PowerToys include TweakUI, a program that lets you make adjustments to more Windows settings than you knew existed. ■ Resource Kits—A set of books published by Microsoft for some versions of Windows that includes a CD-ROM containing hundreds of utility programs. What you may not have known is that in some cases you can download the Resource Kit program toolkits with- out purchasing the books. ■ Subsystem for UNIX-Based Applications (SUA)—A package of network services and command-line tools that provide a nearly complete UNIX environment. It can be installed only on Windows Vista Ultimate and Enterprise, and Windows Server 2003.