Downloaded from Uniprot ( Before Eluting in Rnase-Free Water
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases
G C A T T A C G G C A T genes Review RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases Amber Willbanks, Shaun Wood and Jason X. Cheng * Department of Pathology, Hematopathology Section, University of Chicago, Chicago, IL 60637, USA; [email protected] (A.W.); [email protected] (S.W.) * Correspondence: [email protected] Abstract: Chromatin structure plays an essential role in eukaryotic gene expression and cell identity. Traditionally, DNA and histone modifications have been the focus of chromatin regulation; however, recent molecular and imaging studies have revealed an intimate connection between RNA epigenetics and chromatin structure. Accumulating evidence suggests that RNA serves as the interplay between chromatin and the transcription and splicing machineries within the cell. Additionally, epigenetic modifications of nascent RNAs fine-tune these interactions to regulate gene expression at the co- and post-transcriptional levels in normal cell development and human diseases. This review will provide an overview of recent advances in the emerging field of RNA epigenetics, specifically the role of RNA modifications and RNA modifying proteins in chromatin remodeling, transcription activation and RNA processing, as well as translational implications in human diseases. Keywords: 5’ cap (5’ cap); 7-methylguanosine (m7G); R-loops; N6-methyladenosine (m6A); RNA editing; A-to-I; C-to-U; 2’-O-methylation (Nm); 5-methylcytosine (m5C); NOL1/NOP2/sun domain Citation: Willbanks, A.; Wood, S.; (NSUN); MYC Cheng, J.X. RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases. Genes 2021, 12, 627. -

Structural and Functional Coordination of DNA and Histone Methylation
Structural and Functional Coordination of DNA and Histone Methylation Xiaodong Cheng Department of Biochemistry, Emory University School of Medicine, Atlanta, Georgia 30322 Correspondence: [email protected] SUMMARY One of the most fundamental questions in the control of gene expression in mammals is how epigenetic methylation patterns of DNA and histones are established, erased, and recognized. This central process in controlling gene expression includes coordinated covalent modifications of DNA and its associated histones. This article focuses on structural aspects of enzymatic activities of histone (arginine and lysine) methylation and demethylation and functional links between the methylation status of the DNA and histones. An interconnected network of methyltransferases, demethylases, and accessory proteins is responsible for changing or maintaining the modification status of specific regions of chromatin. Outline 1 Histone methylation and demethylation 4 Summary 2 DNA methylation References 3 Interplay between DNA methylation and histone modification Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg Additional Perspectives on Epigenetics available at www.cshperspectives.org Copyright # 2014 Cold Spring Harbor Laboratory Press; all rights reserved; doi: 10.1101/cshperspect.a018747 Cite this article as Cold Spring Harb Perspect Biol 2014;6:a018747 1 X. Cheng OVERVIEW All cells face the problem of controlling the amounts and articles by Becker and Workman 2013; Allis et al. 2014; timing of expression of their -

Fruit Ripening and Storage
OPEN Citation: Horticulture Research (2014) 1, 6; doi:10.1038/hortres.2014.6 ß 2014 Nanjing Agricultural University All rights reserved 2052-7276/14 www.nature.com/hortres ARTICLE Dynamic changes in proteins during apple (Malus x domestica) fruit ripening and storage Yun Shi1, Li Jiang1, Li Zhang2, Ruoyi Kang1 and Zhifang Yu1 A proteomic study, using two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight/time-of-flight, was conducted in apple fruit (cv. ‘Golden Delicious’) starting at 10 days prior to harvest through 50 days in storage. Total protein was extracted using a phenol/sodium dodecyl sulfate protocol. More than 400 protein spots were detected in each gel and 55 differentially expressed proteins (p,0.05) were subjected to matrix-assisted laser desorption/ionization time-of-flight/ time-of-flight analysis. Fifty-three of these proteins were finally identified using an apple expressed sequence tag database downloaded from Genome Database for Rosaceae and placed into six categories. The categories and the percentage of proteins placed in each category were stress response and defense (49.0%), energy and metabolism (34.0%), fruit ripening and senescence (5.6%), signal transduction (3.8%), cell structure (3.8%) and protein synthesis (3.8%). Proteins involved in several multiple metabolic pathways, including glycolysis, pentose–phosphate pathway, anti-oxidative systems, photosynthesis and cell wall synthesis, were downregulated, especially during the climacteric burst in respiration and during the senescent stages of fruit development. Proteins classified as allergens or involved in cell wall degradation were upregulated during the ripening process. Some protein spots exhibited a mixed pattern (increasing to maximal abundance followed by a decrease), such as 1-aminocyclopropane-1-carboxylate oxidase, L-ascorbate peroxidase and abscisic acid response proteins. -

Genome-Wide Analysis of Glyoxalase-Like Gene Families in Grape
Li et al. BMC Genomics (2019) 20:362 https://doi.org/10.1186/s12864-019-5733-y RESEARCHARTICLE Open Access Genome-wide analysis of glyoxalase-like gene families in grape (Vitis vinifera L.) and their expression profiling in response to downy mildew infection Tiemei Li1,2,3, Xin Cheng1,2,3, Yuting Wang1,2,3, Xiao Yin1,2,3, Zhiqian Li1,2,3, Ruiqi Liu1,2,3, Guotian Liu1,2,3, Yuejin Wang1,2,3 and Yan Xu1,2,3* Abstract Background: The glyoxalase system usually comprises two enzymes, glyoxalase I (GLYI) and glyoxalase II (GLYII). This system converts cytotoxic methylglyoxal (MG) into non-toxic D-lactate in the presence of reduced glutathione (GSH) in two enzymatic steps. Recently, a novel type of glyoxalase III (GLYIII) activity has observed in Escherichia coli that can detoxify MG into D-lactate directly, in one step, without a cofactor. Investigation of the glyoxalase enzymes of a number of plant species shows the importance of their roles in response both to abiotic and to biotic stresses. Until now, glyoxalase gene families have been identified in the genomes of four plants, Arabidopsis, Oryza sativa, Glycine max and Medicago truncatula but no similar study has been done with the grapevine Vitis vinifera L. Results: In this study, four GLYI-like,twoGLYII-like and three GLYIII-like genesareidentifiedfromthegenomedatabaseof grape. All these genes were analysed in detail, including their chromosomal locations, phylogenetic relationships, exon-intron distributions, protein domain organisations and the presence of conserved binding sites. Using quantitative real-time PCR analysis (qRT-PCR), the expression profiles of these geneswereanalysedindifferent tissues of grape, and also when under infection stress from downy mildew (Plasmopara viticola). -

From 1957 to Nowadays: a Brief History of Epigenetics
International Journal of Molecular Sciences Review From 1957 to Nowadays: A Brief History of Epigenetics Paul Peixoto 1,2, Pierre-François Cartron 3,4,5,6,7,8, Aurélien A. Serandour 3,4,6,7,8 and Eric Hervouet 1,2,9,* 1 Univ. Bourgogne Franche-Comté, INSERM, EFS BFC, UMR1098, Interactions Hôte-Greffon-Tumeur/Ingénierie Cellulaire et Génique, F-25000 Besançon, France; [email protected] 2 EPIGENEXP Platform, Univ. Bourgogne Franche-Comté, F-25000 Besançon, France 3 CRCINA, INSERM, Université de Nantes, 44000 Nantes, France; [email protected] (P.-F.C.); [email protected] (A.A.S.) 4 Equipe Apoptose et Progression Tumorale, LaBCT, Institut de Cancérologie de l’Ouest, 44805 Saint Herblain, France 5 Cancéropole Grand-Ouest, Réseau Niches et Epigénétique des Tumeurs (NET), 44000 Nantes, France 6 EpiSAVMEN Network (Région Pays de la Loire), 44000 Nantes, France 7 LabEX IGO, Université de Nantes, 44000 Nantes, France 8 Ecole Centrale Nantes, 44300 Nantes, France 9 DImaCell Platform, Univ. Bourgogne Franche-Comté, F-25000 Besançon, France * Correspondence: [email protected] Received: 9 September 2020; Accepted: 13 October 2020; Published: 14 October 2020 Abstract: Due to the spectacular number of studies focusing on epigenetics in the last few decades, and particularly for the last few years, the availability of a chronology of epigenetics appears essential. Indeed, our review places epigenetic events and the identification of the main epigenetic writers, readers and erasers on a historic scale. This review helps to understand the increasing knowledge in molecular and cellular biology, the development of new biochemical techniques and advances in epigenetics and, more importantly, the roles played by epigenetics in many physiological and pathological situations. -

NIH Public Access Provided by CDC Stacks Author Manuscript Mol Genet Metab
View metadata, citation and similar papers at core.ac.uk brought to you by CORE NIH Public Access provided by CDC Stacks Author Manuscript Mol Genet Metab. Author manuscript; available in PMC 2013 November 01. Published in final edited form as: Mol Genet Metab. 2012 November ; 107(3): 596–604. doi:10.1016/j.ymgme.2012.09.022. Associations between maternal genotypes and metabolites implicated in congenital heart defects $watermark-text $watermark-text $watermark-text Shimul Chowdhury1,2, Charlotte A. Hobbs1, Stewart L. MacLeod1, Mario A. Cleves1, Stepan Melnyk1, S. Jill James1, Ping Hu1, and Stephen W. Erickson1,3 1Department of Pediatrics, College of Medicine, University of Arkansas for Medical Sciences, Arkansas Children’s Hospital Research Institute, 13 Children’s Way, Slot 512, Little Rock, AR 72202, USA 2Clinical Molecular Genetics Department, Providence Sacred Heart Medical Center, 101 W. Eighth Avenue, Spokane, WA 99204, USA 3Department of Biostatistics, College of Medicine, University of Arkansas for Medical Sciences, Arkansas Children’s Hospital Research Institute, 4301 W. Markham Street, Slot 781, Little Rock, AR 72205, USA Abstract Background—The development of non-syndromic congenital heart defects (CHDs) involves a complex interplay of genetics, metabolism, and lifestyle. Previous studies have implicated maternal single nucleotide polymorphisms (SNPs) and altered metabolism in folate-related pathways as CHD risk factors. Objective—We sought to discover associations between maternal SNPs and metabolites involved in the homocysteine, folate, and transsulfuration pathways, and determine if these associations differ between CHD cases and controls. Design—Genetic, metabolic, demographic, and lifestyle information was available for 335 mothers with CHD-affected pregnancies and 263 mothers with unaffected pregnancies. -
Supplementary File 1
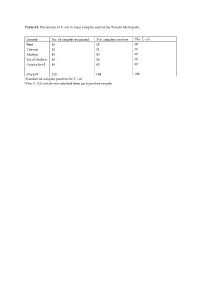
Table S1. Prevalence of E. coli in meat samples sold at the Tamale Metropolis. Sample No. of samples examined aNo. samples positive bNo. E. coli Beef 45 39 39 Chevon 45 34 34 Mutton 45 40 40 Local chicken 45 36 36 Guinea fowl 45 40 40 Overall 225 189 189 aNumber of samples positive for E. coli. bOne E. Coli isolate was selected from each positive sample. Table S2. A table showing the eBURST (Based Upon Related Sequence Types) analyses of the study sequence types with global curated STs in Escherichia PubMLST database. MLST (Isolate) Type of clone Closet global ancestry Source sequence type (ST) ST69 (SG6) Similar a ST69 Animal (Food), Human ST155 (SLC2, Similar ST155 Animal (Food), Human, TLC13, CM4) Environment ST297 (TLC1) Similar ST297 Human ST1727 (NC3) Similar ST1727 Human ST44 (AC1) Single-Locus Variant ST10, ST752 Animal (Food), (SLV) b Human ST469 (CC6) Single-Locus Variant ST162 Food (SLV) ST540 (AB1, Single-Locus Variant ST4093 Human TG1) (SLV) ST1141 (NM11) Single-Locus Variant ST10, ST744 Animal (Food), (SLV) Human ST7473 (NB12) Single-Locus Variant ST10 Animal (Food), (SLV) Human ST6646 (CB1) Satellite c None - ST7483 (NB12) Satellite None - a Similar: study isolate was similar to a global curated known sequence type. b Single-Locus Variant (SLV): study isolate only shared similarity with global curated known sequence types that differed in one allelic gene. c Satellite: study isolate as a distantly related and did not shared any similarity with global curated known sequence types. Table S3. In silico identification and characterization of conserved stress response mechanisms in the E. -

The Role of B-Vitamins – Gene Interactions in Colorectal Carcinogenesis a Molecular Epidemiological Approach
THE ROLE OF B-VITAMINS – GENE INTERACTIONS IN COLORECTAL CARCINOGENESIS A MOLECULAR EPIDEMIOLOGICAL APPROACH Promotor Prof. dr. ir. F.J. Kok Hoogleraar Voeding en Gezondheid Wageningen Universiteit Co-promotoren Dr. ir. E. Kampman Universitair Hoofddocent, sectie Humane Voeding Wageningen Universiteit Dr. J. Keijer Hoofd Food Bioactives Group RIKILT – Instituut voor Voedselveiligheid Promotiecommissie Prof. dr. J. Mathers University of Newcastle Upon Tyne, United Kingdom Prof. dr. G.A. Meijer VU Medisch Centrum, Amsterdam Dr. ir. P. Verhoef Wageningen Centre for Food Sciences Prof. dr. S.C. de Vries Wageningen Universiteit Dit onderzoek is uitgevoerd binnen de onderzoeksschool VLAG. THE ROLE OF B-VITAMINS – GENE INTERACTIONS IN COLORECTAL CARCINOGENESIS A MOLECULAR EPIDEMIOLOGICAL APPROACH Maureen van den Donk Proefschrift ter verkrijging van de graad van doctor op gezag van de rector magnificus van Wageningen Universiteit, Prof. dr. M.J. Kropff, in het openbaar te verdedigen op dinsdag 13 december 2005 des namiddags te half twee in de Aula Maureen van den Donk The role of B-vitamins – gene interactions in colorectal carcinogenesis – A molecular epidemiological approach Thesis Wageningen University – With references – With summary in Dutch ISBN 90-8504-328-X ABSTRACT The role of B-vitamins–gene interactions in colorectal carcinogenesis – A molecular epidemiological approach PhD thesis by Maureen van den Donk, Division of Human Nutrition, Wageningen University, The Netherlands, December 13, 2005. Folate deficiency can affect DNA methylation and DNA synthesis. Both factors may be operative in colorectal carcinogenesis. Many enzymes, like methylenetetrahydrofolate reductase (MTHFR), thymidylate synthase (TS), methionine synthase (MTR), and serine hydroxymethyltransferase (SHMT), are needed for conversions in folate metabolism. -

Supplementary Information 2 to Accompany
1 Supplementary Information 2 to accompany 3 Sulfur-oxidizing symbionts without canonical genes for autotrophic CO2 fixation 4 Brandon K. B. Seah*1,7, Chakkiath Paul Antony1,8, Bruno Huettel2, Jan Zarzycki3, Lennart 5 Schada von Borzyskowski3, Tobias J. Erb3, Angela Kouris4, Manuel Kleiner5, Manuel 6 Liebeke1, Nicole Dubilier1,6, Harald R. Gruber-Vodicka1 7 1 Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany 8 2 Max Planck Genome Centre Cologne, Max Planck Institute for Plant Breeding Research, 9 Carl-von-Linné-Weg 10, 50829 Cologne, Germany 10 3 Max Planck Institute for Terrestrial Microbiology, Karl-von-Frisch-Str. 10, 35043 Marburg, 11 Germany 12 4 Energy Bioengineering and Geomicrobiology Group, University of Calgary, 2500 13 University Drive Northwest, Calgary, Alberta T2N 1N4, Canada 14 5 Department of Plant and Microbial Biology, North Carolina State University, Raleigh 15 27695, North Carolina, United States of America 16 6 MARUM, Center for Marine Environmental Sciences, University of Bremen, 28359 17 Bremen, Germany 18 7 Current address: Max Planck Institute for Developmental Biology, Max-Planck-Ring 5, 19 72076 Tübingen, Germany 20 8 Current address: Red Sea Research Center, Biological and Environmental Sciences and 21 Engineering (BESE) Division, King Abdullah University of Science and Technology 22 (KAUST), Thuwal 23955, Kingdom of Saudi Arabia 23 * Corresponding author 1 24 Supplementary Materials and Methods 25 Metabolite extraction and identification 26 Kentrophoros sp. H was collected on Elba in 2014 for metabolomics (Supplementary Table 27 7). Samples were fixed in 1 mL cold methanol (HPLC-grade, Sigma-Aldrich) and stored at - 28 20°C until use. -

Overview of Distinct 5-Methylcytosine Profiles of Messenger RNA In
Zhang et al. J Transl Med (2020) 18:245 https://doi.org/10.1186/s12967-020-02417-6 Journal of Translational Medicine RESEARCH Open Access Overview of distinct 5-methylcytosine profles of messenger RNA in human hepatocellular carcinoma and paired adjacent non-tumor tissues Qiyao Zhang1,2,3,4†, Qingyuan Zheng1,2,3,4†, Xiao Yu1,2,3,4†, Yuting He1,2,3,4* and Wenzhi Guo1,2,3,4* Abstract Background: Post-transcriptional methylation modifcations, including 5-methylcytosine (m5C) modifcation, are closely related to the tumorigenesis of cancers. However, the mRNA profle of m5C modifcation in hepatocellular carcinoma (HCC) is unknown. Methods: Methylated RNA immunoprecipitation sequencing was performed to identify m5C peaks on mRNA of human HCC tissues and adjacent tissues, and diferences in m5C between the two groups were analyzed. In addition, we conducted a bioinformatics analysis to predict the function of specifc methylated transcripts. Results: We found that there was a noticeable diference in m5C between HCC and paired non-tumor tissues, sug- gesting that m5C could play a role in the pathogenesis of HCC. In addition, analyses of gene ontology and the Kyoto Encyclopedia of Genes and Genomes showed that the unique distribution pattern of mRNA m5C in HCC was associ- ated with a wide range of cellular functions. Conclusions: Our results revealed diferent distribution patterns of m5C in HCC and adjacent tissues and provided new insights into a novel function of m5C RNA methylation of mRNA in HCC progression. Keywords: mRNA, 5-methylcytosine, Hepatocellular carcinoma, RNA methylation, MeRIP-seq Background treatment, due to its late diagnosis, high metastasis, and Hepatocellular carcinoma (HCC) is one of the most high recurrence rate, the lethal rate of hepatocellular widespread cancers, and it has an extremely poor prog- carcinoma remains high [4–7]. -

Stress-Related Methylation of the Catechol-O-Methyltransferase Val158 Allele Predicts Human Prefrontal Cognition and Activity
6692 • The Journal of Neuroscience, May 4, 2011 • 31(18):6692–6698 Behavioral/Systems/Cognitive Stress-Related Methylation of the Catechol-O-Methyltransferase Val158 Allele Predicts Human Prefrontal Cognition and Activity Gianluca Ursini,1,2 Valentina Bollati,3,4 Leonardo Fazio,1 Annamaria Porcelli,1 Luisa Iacovelli,5 Assia Catalani,5 Lorenzo Sinibaldi,2 Barbara Gelao,1 Raffaella Romano,1 Antonio Rampino,1 Paolo Taurisano,1 Marina Mancini,1 Annabella Di Giorgio,1,6 Teresa Popolizio,6 Andrea Baccarelli,3,4,7 Antonio De Blasi,8 Giuseppe Blasi,1 and Alessandro Bertolino1,6 1Psychiatric Neuroscience Group, Department of Neurological and Psychiatric Sciences, University “Aldo Moro”, 71024 Bari, Italy, 2Mendel Laboratory, Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) “Casa Sollievo della Sofferenza”, 71013 San Giovanni Rotondo, Italy, 3Center for Molecular and Genetic Epidemiology, Department of Environmental and Occupational Health, University of Milan, Milan, Italy, 4Istituto Di Ricovero e Cura a Carattere Scientifico Maggiore Hospital, Mangiagalli and Regina Elena Foundation, 20122 Milan, Italy, 5Department of Physiology and Pharmacology “V. Erspamer”, University of Rome “Sapienza”, 00185 Rome, Italy, 6Department of Neuroradiology, IRCCS “Casa Sollievo della Sofferenza”, 71013 San Giovanni Rotondo, Italy, 7Exposure, Epidemiology, and Risk Program, Department of Environmental Health, Harvard School of Public Health, Boston, Massachusetts 02115- 6018, and 8Department of Molecular Medicine, University of Rome “Sapienza”, 00185 Rome, Italy DNA methylation at CpG dinucleotides is associated with gene silencing, stress, and memory. The catechol-O-methyltransferase (COMT) Val 158 allele in rs4680 is associated with differential enzyme activity, stress responsivity, and prefrontal activity during working memory (WM), and it creates a CpG dinucleotide. -

Recombinant Human Glyoxalase I Catalog Number: 4959-GL
Recombinant Human Glyoxalase I Catalog Number: 4959-GL DESCRIPTION Source E. coliderived Ala2Met184, with an Nterminal Met and 6His tag Accession # NP_006699 Nterminal Sequence Met Analysis Predicted Molecular 22 kDa Mass SPECIFICATIONS SDSPAGE 25 kDa, reducing conditions Activity Measured by its ability to catalyze the formation of SDlactoylglutathione from the hemimercaptal adduct that forms spontaneously between methylglyoxal and reduced glutathione. The specific activity is >100 nmol/min/µg, as measured under the described conditions. Endotoxin Level <1.0 EU per 1 μg of the protein by the LAL method. Purity >85%, by SDSPAGE under reducing conditions and visualized by silver stain. Formulation Lyophilized from a 0.2 μm filtered solution in TrisHCl and DTT. See Certificate of Analysis for details. Activity Assay Protocol Materials l Assay Buffer: 0.1 M Sodium Phosphate, pH 7.0 l Recombinant Human Glyoxalase I (rhGlyoxalase I) (Catalog # 4959GL) l Glutathione, Reduced (GSH) (Amresco, Catalog # 0399) l Methylglyoxal solution, 40% (Sigma, Catalog # M0252) l 96well Clear UV Plate (Costar, Catalog # 3635) l Plate Reader (Model: SpectraMax Plus by Molecular Devices) or equivalent Assay 1. Prepare 100 mM GSH in deionized water. Note: Prepare fresh. 2. Dilute 40% (6.48 M) Methylglyoxal solution to 100 mM in Assay Buffer. Note: Prepare fresh. 3. Combine 1420 µL Assay Buffer, 40 µL 100 mM GSH, and 40 µL 100 mM Methylglyoxal to make the Substrate Mixture. 4. Incubate at room temperature for 15 minutes. 5. Dilute rhGlyoxalase I to 0.4 ng/µL in Assay Buffer.