Mobilization of Iron Stored in Bacterioferritin Is Required for Metabolic Homeostasis in Pseudomonas Aeruginosa
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional Properties and Molecular Architecture of Leukotriene A4 Hydrolase, a Pivotal Catalyst of Chemotactic Leukotriene Formation
Review Article TheScientificWorldJOURNAL (2002) 2, 1734–1749 ISSN 1537-744X; DOI 10.1100/tsw.2002.810 Functional Properties and Molecular Architecture of Leukotriene A4 Hydrolase, a Pivotal Catalyst of Chemotactic Leukotriene Formation Jesper Z. Haeggström1,*, Pär Nordlund2, and Marjolein M.G.M. Thunnissen2 1Department of Medical Biochemistry and Biophysics, Division of Chemistry 2, Karolinska Institutet, S-171 77 Stockholm, Sweden; 2Department of Biochemistry, University of Stockholm, Arrhenius Laboratories A4, S-106 91 Stockholm, Sweden E-mail: [email protected]; [email protected]; [email protected] Received March 25, 2002; Accepted April 26, 2002; Published June 26, 2002 The leukotrienes are a family of lipid mediators involved in inflammation and allergy. Leukotriene B4 is a classical chemoattractant, which triggers adherence and aggregation of leukocytes to the endothelium at only nM concentrations. In addition, leukotriene B4 modulates immune responses, participates in the host defense against infections, and is a key mediator of PAF-induced lethal shock. Because of these powerful biological effects, leukotriene B4 is implicated in a variety of acute and chronic inflammatory diseases, e.g., nephritis, arthritis, dermatitis, and chronic obstructive pulmonary disease. The final step in the biosynthesis of leukotriene B4 is catalyzed by leukotriene A4 hydrolase, a unique bifunctional zinc metalloenzyme with an anion-dependent aminopeptidase activity. Here we describe the most recent developments regarding our understanding -

Role of Epoxide Hydrolases in Lipid Metabolism
Biochimie 95 (2013) 91e95 Contents lists available at SciVerse ScienceDirect Biochimie journal homepage: www.elsevier.com/locate/biochi Mini-review Role of epoxide hydrolases in lipid metabolism Christophe Morisseau* Department of Entomology and U.C.D. Comprehensive Cancer Center, One Shields Avenue, University of California, Davis, CA 95616, USA article info abstract Article history: Epoxide hydrolases (EH), enzymes present in all living organisms, transform epoxide-containing lipids to Received 29 March 2012 1,2-diols by the addition of a molecule of water. Many of these oxygenated lipid substrates have potent Accepted 8 June 2012 biological activities: host defense, control of development, regulation of blood pressure, inflammation, Available online 18 June 2012 and pain. In general, the bioactivity of these natural epoxides is significantly reduced upon metabolism to diols. Thus, through the regulation of the titer of lipid epoxides, EHs have important and diverse bio- Keywords: logical roles with profound effects on the physiological state of the host organism. This review will Epoxide hydrolase discuss the biological activity of key lipid epoxides in mammals. In addition, the use of EH specific Epoxy-fatty acids Cholesterol epoxide inhibitors will be highlighted as possible therapeutic disease interventions. Ó Juvenile hormone 2012 Elsevier Masson SAS. All rights reserved. 1. Introduction hydrolyzed by a water molecule [8]. Based on this mechanism, transition-state inhibitors of EHs have been designed (Fig. 1B). Epoxides are three atom cyclic ethers formed by the oxidation of These ureas and amides are tight-binding competitive inhibitors olefins. Because of their highly polarized oxygen-carbon bonds and with low nanomolar dissociation constants (KI) [9] [10]. -

Quantum Chemical Studies of Epoxide- Transforming Enzymes
Quantum Chemical Studies of Epoxide- Transforming Enzymes Kathrin H. Hopmann Department of Theoretical Chemistry Royal Institute of Technology Stockholm, Sweden, 2007 ii © Kathrin H. Hopmann, 2007 ISBN 978-91-7178-640-1 ISSN 1654-2312 TRITA-BIO-Report 2007:3 Printed by Universitetsservice US-AB, Stockholm, Sweden. iii Abstract Density functional theory is employed to study the reaction mechanisms of different epoxide-transforming enzymes. Calculations are based on quantum chemical active site models, which are build from X-ray crystal structures. The models are used to study conversion of various epoxides into their corresponding diols or substituted alcohols. Epoxide-transforming enzymes from three different families are studied. The human soluble epoxide hydrolase (sEH) belongs to the α/β-hydrolase fold family. sEH employs a covalent mechanism to hydrolyze various epoxides into vicinal diols. The Rhodococcus erythrobacter limonene epoxide hydrolase (LEH) constitutes a novel epoxide hydrolase, which is considered the founding member of a new family of enzymes. LEH mediates transformation of limone-1,2-epoxide into the corresponding vicinal diol by employing a general acid/general base-mediated mechanism. The Agrobacterium radiobacter AD1 haloalcohol dehalogenase HheC is related to the short-chain dehydrogenase/reductases. HheC is able to convert epoxides using various nucleophiles such as azide, cyanide, and nitrite. Reaction mechanisms of these three enzymes are analyzed in depth and the role of different active site residues is studied through in silico mutations. Steric and electronic factors influencing the regioselectivity of epoxide opening are identified. The computed energetics help to explain preferred reaction pathways and experimentally observed regioselectivities. Our results confirm the usefulness of the employed computational methodology for investigating enzymatic reactions. -

1 Metabolic Dysfunction Is Restricted to the Sciatic Nerve in Experimental
Page 1 of 255 Diabetes Metabolic dysfunction is restricted to the sciatic nerve in experimental diabetic neuropathy Oliver J. Freeman1,2, Richard D. Unwin2,3, Andrew W. Dowsey2,3, Paul Begley2,3, Sumia Ali1, Katherine A. Hollywood2,3, Nitin Rustogi2,3, Rasmus S. Petersen1, Warwick B. Dunn2,3†, Garth J.S. Cooper2,3,4,5* & Natalie J. Gardiner1* 1 Faculty of Life Sciences, University of Manchester, UK 2 Centre for Advanced Discovery and Experimental Therapeutics (CADET), Central Manchester University Hospitals NHS Foundation Trust, Manchester Academic Health Sciences Centre, Manchester, UK 3 Centre for Endocrinology and Diabetes, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, UK 4 School of Biological Sciences, University of Auckland, New Zealand 5 Department of Pharmacology, Medical Sciences Division, University of Oxford, UK † Present address: School of Biosciences, University of Birmingham, UK *Joint corresponding authors: Natalie J. Gardiner and Garth J.S. Cooper Email: [email protected]; [email protected] Address: University of Manchester, AV Hill Building, Oxford Road, Manchester, M13 9PT, United Kingdom Telephone: +44 161 275 5768; +44 161 701 0240 Word count: 4,490 Number of tables: 1, Number of figures: 6 Running title: Metabolic dysfunction in diabetic neuropathy 1 Diabetes Publish Ahead of Print, published online October 15, 2015 Diabetes Page 2 of 255 Abstract High glucose levels in the peripheral nervous system (PNS) have been implicated in the pathogenesis of diabetic neuropathy (DN). However our understanding of the molecular mechanisms which cause the marked distal pathology is incomplete. Here we performed a comprehensive, system-wide analysis of the PNS of a rodent model of DN. -

Mechanistic Implications for the Chorismatase Fkbo Based on the Crystal Structure
Mechanistic Implications for the Chorismatase FkbO Based on the Crystal Structure Puneet Juneja 1,†, Florian Hubrich 2,†, Kay Diederichs 1, Wolfram Welte 1 and Jennifer N. Andexer 2 1 - Department of Biology, University of Konstanz, D-78457 Konstanz, Germany 2 - Institute of Pharmaceutical Sciences, Albert-Ludwigs-University Freiburg, Albertstr 25, D-79104 Freiburg, Germany Correspondence to Jennifer N. Andexer: [email protected] http://dx.doi.org/10.1016/j.jmb.2013.09.006 Edited by M. Guss Abstract Chorismate-converting enzymes are involved in many biosynthetic pathways leading to natural products and can often be used as tools for the synthesis of chemical building blocks. Chorismatases such as FkbO from Streptomyces species catalyse the hydrolysis of chorismate yielding (dihydro)benzoic acid derivatives. In contrast to many other chorismate-converting enzymes, the structure and catalytic mechanism of a chorismatase had not been previously elucidated. Here we present the crystal structure of the chorismatase FkbO in complex with a competitive inhibitor at 1.08 Å resolution. FkbO is a monomer in solution and exhibits pseudo-3-fold symmetry; the structure of the individual domains indicates a possible connection to the trimeric RidA/YjgF family and related enzymes. The co-crystallised inhibitor led to the identification of FkbO's active site in the cleft between the central and the C-terminal domains. A mechanism for FkbO is proposed based on both interactions between the inhibitor and the surrounding amino acids and an FkbO structure with chorismate modelled in the active site. We suggest that the methylene group of the chorismate enol ether takes up a proton from an active-site glutamic acid residue, thereby initiating chorismate hydrolysis. -

Fruit Ripening and Storage
OPEN Citation: Horticulture Research (2014) 1, 6; doi:10.1038/hortres.2014.6 ß 2014 Nanjing Agricultural University All rights reserved 2052-7276/14 www.nature.com/hortres ARTICLE Dynamic changes in proteins during apple (Malus x domestica) fruit ripening and storage Yun Shi1, Li Jiang1, Li Zhang2, Ruoyi Kang1 and Zhifang Yu1 A proteomic study, using two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight/time-of-flight, was conducted in apple fruit (cv. ‘Golden Delicious’) starting at 10 days prior to harvest through 50 days in storage. Total protein was extracted using a phenol/sodium dodecyl sulfate protocol. More than 400 protein spots were detected in each gel and 55 differentially expressed proteins (p,0.05) were subjected to matrix-assisted laser desorption/ionization time-of-flight/ time-of-flight analysis. Fifty-three of these proteins were finally identified using an apple expressed sequence tag database downloaded from Genome Database for Rosaceae and placed into six categories. The categories and the percentage of proteins placed in each category were stress response and defense (49.0%), energy and metabolism (34.0%), fruit ripening and senescence (5.6%), signal transduction (3.8%), cell structure (3.8%) and protein synthesis (3.8%). Proteins involved in several multiple metabolic pathways, including glycolysis, pentose–phosphate pathway, anti-oxidative systems, photosynthesis and cell wall synthesis, were downregulated, especially during the climacteric burst in respiration and during the senescent stages of fruit development. Proteins classified as allergens or involved in cell wall degradation were upregulated during the ripening process. Some protein spots exhibited a mixed pattern (increasing to maximal abundance followed by a decrease), such as 1-aminocyclopropane-1-carboxylate oxidase, L-ascorbate peroxidase and abscisic acid response proteins. -

Genome-Wide Analysis of Glyoxalase-Like Gene Families in Grape
Li et al. BMC Genomics (2019) 20:362 https://doi.org/10.1186/s12864-019-5733-y RESEARCHARTICLE Open Access Genome-wide analysis of glyoxalase-like gene families in grape (Vitis vinifera L.) and their expression profiling in response to downy mildew infection Tiemei Li1,2,3, Xin Cheng1,2,3, Yuting Wang1,2,3, Xiao Yin1,2,3, Zhiqian Li1,2,3, Ruiqi Liu1,2,3, Guotian Liu1,2,3, Yuejin Wang1,2,3 and Yan Xu1,2,3* Abstract Background: The glyoxalase system usually comprises two enzymes, glyoxalase I (GLYI) and glyoxalase II (GLYII). This system converts cytotoxic methylglyoxal (MG) into non-toxic D-lactate in the presence of reduced glutathione (GSH) in two enzymatic steps. Recently, a novel type of glyoxalase III (GLYIII) activity has observed in Escherichia coli that can detoxify MG into D-lactate directly, in one step, without a cofactor. Investigation of the glyoxalase enzymes of a number of plant species shows the importance of their roles in response both to abiotic and to biotic stresses. Until now, glyoxalase gene families have been identified in the genomes of four plants, Arabidopsis, Oryza sativa, Glycine max and Medicago truncatula but no similar study has been done with the grapevine Vitis vinifera L. Results: In this study, four GLYI-like,twoGLYII-like and three GLYIII-like genesareidentifiedfromthegenomedatabaseof grape. All these genes were analysed in detail, including their chromosomal locations, phylogenetic relationships, exon-intron distributions, protein domain organisations and the presence of conserved binding sites. Using quantitative real-time PCR analysis (qRT-PCR), the expression profiles of these geneswereanalysedindifferent tissues of grape, and also when under infection stress from downy mildew (Plasmopara viticola). -

Download (2MB)
Enzyme and Microbial Technology 139 (2020) 109592 Contents lists available at ScienceDirect Enzyme and Microbial Technology journal homepage: www.elsevier.com/locate/enzmictec Identification and catalytic properties of new epoxide hydrolases fromthe T genomic data of soil bacteria Gorjan Stojanovskia, Dragana Dobrijevica, Helen C. Hailesb, John M. Warda,* a Department of Biochemical Engineering, University College London, Bernard Katz, London WC1E 6BT, UK b Department of Chemistry, University College London, 20 Gordon Street, London, WC1H 0AJ, UK ARTICLE INFO ABSTRACT Keywords: Epoxide hydrolases (EHs) catalyse the conversion of epoxides into vicinal diols. These enzymes have extensive Epoxide hydrolase value in biocatalysis as they can generate enantiopure epoxides and diols which are important and versatile Limonene epoxide hydrolase synthetic intermediates for the fine chemical and pharmaceutical industries. Despite these benefits, theyhave Genome mining seen limited use in the bioindustry and novel EHs continue to be reported in the literature. Biotransformation We identified twenty-nine putative EHs within the genomes of soil bacteria. Eight of these EHs wereexplored in terms of their activity. Two limonene epoxide hydrolases (LEHs) and one ⍺/β EH were active on a model compound styrene oxide and its ring-substituted derivatives, with low to good percentage conversions of 18–86%. Further exploration of the substrate scope with enantiopure (R)-styrene oxide and (S)-styrene oxide, showed different epoxide ring opening regioselectivities. Two enzymes, expressed from plasmids pQR1984and pQR1990 de-symmetrised the meso-epoxide cyclohexene oxide, forming the (R,R)-diol with high enantioselec- tivity. Two LEHs, from plasmids pQR1980 and pQR1982 catalysed the hydrolysis of (+) and (−) limonene oxide, with diastereomeric preference for the (1S,2S,4R)- and (1R,2R,4S)-diol products, respectively. -

Phza/B Catalyzes the Formation of the Tricycle in Phenazine Biosynthesis Ekta G
Subscriber access provided by DigiTop | USDA's Digital Desktop Library Article PhzA/B Catalyzes the Formation of the Tricycle in Phenazine Biosynthesis Ekta G. Ahuja, Petra Janning, Matthias Mentel, Almut Graebsch, Rolf Breinbauer, Wolf Hiller, Burkhard Costisella, Linda S. Thomashow, Dmitri V. Mavrodi, and Wulf Blankenfeldt J. Am. Chem. Soc., 2008, 130 (50), 17053-17061 • DOI: 10.1021/ja806325k • Publication Date (Web): 17 November 2008 Downloaded from http://pubs.acs.org on January 15, 2009 More About This Article Additional resources and features associated with this article are available within the HTML version: • Supporting Information • Access to high resolution figures • Links to articles and content related to this article • Copyright permission to reproduce figures and/or text from this article Journal of the American Chemical Society is published by the American Chemical Society. 1155 Sixteenth Street N.W., Washington, DC 20036 Published on Web 11/17/2008 PhzA/B Catalyzes the Formation of the Tricycle in Phenazine Biosynthesis Ekta G. Ahuja,† Petra Janning,† Matthias Mentel,‡,§ Almut Graebsch,‡ Rolf Breinbauer,†,‡,§,| Wolf Hiller,‡ Burkhard Costisella,‡ Linda S. Thomashow,⊥,# Dmitri V. Mavrodi,⊥ and Wulf Blankenfeldt*,† Max-Planck-Institute of Molecular Physiology, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany, Technical UniVersity of Dortmund, Faculty of Chemistry, Otto-Hahn-Strasse 6, 44221 Dortmund, Germany, UniVersity of Leipzig, Institute of Organic Chemistry, Johannisallee 29, 04103 Leipzig, Germany, Graz UniVersity of -
Supplementary File 1
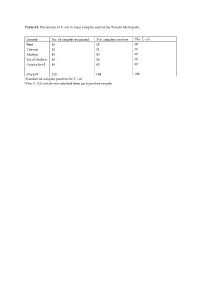
Table S1. Prevalence of E. coli in meat samples sold at the Tamale Metropolis. Sample No. of samples examined aNo. samples positive bNo. E. coli Beef 45 39 39 Chevon 45 34 34 Mutton 45 40 40 Local chicken 45 36 36 Guinea fowl 45 40 40 Overall 225 189 189 aNumber of samples positive for E. coli. bOne E. Coli isolate was selected from each positive sample. Table S2. A table showing the eBURST (Based Upon Related Sequence Types) analyses of the study sequence types with global curated STs in Escherichia PubMLST database. MLST (Isolate) Type of clone Closet global ancestry Source sequence type (ST) ST69 (SG6) Similar a ST69 Animal (Food), Human ST155 (SLC2, Similar ST155 Animal (Food), Human, TLC13, CM4) Environment ST297 (TLC1) Similar ST297 Human ST1727 (NC3) Similar ST1727 Human ST44 (AC1) Single-Locus Variant ST10, ST752 Animal (Food), (SLV) b Human ST469 (CC6) Single-Locus Variant ST162 Food (SLV) ST540 (AB1, Single-Locus Variant ST4093 Human TG1) (SLV) ST1141 (NM11) Single-Locus Variant ST10, ST744 Animal (Food), (SLV) Human ST7473 (NB12) Single-Locus Variant ST10 Animal (Food), (SLV) Human ST6646 (CB1) Satellite c None - ST7483 (NB12) Satellite None - a Similar: study isolate was similar to a global curated known sequence type. b Single-Locus Variant (SLV): study isolate only shared similarity with global curated known sequence types that differed in one allelic gene. c Satellite: study isolate as a distantly related and did not shared any similarity with global curated known sequence types. Table S3. In silico identification and characterization of conserved stress response mechanisms in the E. -

Supplementary Information 2 to Accompany
1 Supplementary Information 2 to accompany 3 Sulfur-oxidizing symbionts without canonical genes for autotrophic CO2 fixation 4 Brandon K. B. Seah*1,7, Chakkiath Paul Antony1,8, Bruno Huettel2, Jan Zarzycki3, Lennart 5 Schada von Borzyskowski3, Tobias J. Erb3, Angela Kouris4, Manuel Kleiner5, Manuel 6 Liebeke1, Nicole Dubilier1,6, Harald R. Gruber-Vodicka1 7 1 Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany 8 2 Max Planck Genome Centre Cologne, Max Planck Institute for Plant Breeding Research, 9 Carl-von-Linné-Weg 10, 50829 Cologne, Germany 10 3 Max Planck Institute for Terrestrial Microbiology, Karl-von-Frisch-Str. 10, 35043 Marburg, 11 Germany 12 4 Energy Bioengineering and Geomicrobiology Group, University of Calgary, 2500 13 University Drive Northwest, Calgary, Alberta T2N 1N4, Canada 14 5 Department of Plant and Microbial Biology, North Carolina State University, Raleigh 15 27695, North Carolina, United States of America 16 6 MARUM, Center for Marine Environmental Sciences, University of Bremen, 28359 17 Bremen, Germany 18 7 Current address: Max Planck Institute for Developmental Biology, Max-Planck-Ring 5, 19 72076 Tübingen, Germany 20 8 Current address: Red Sea Research Center, Biological and Environmental Sciences and 21 Engineering (BESE) Division, King Abdullah University of Science and Technology 22 (KAUST), Thuwal 23955, Kingdom of Saudi Arabia 23 * Corresponding author 1 24 Supplementary Materials and Methods 25 Metabolite extraction and identification 26 Kentrophoros sp. H was collected on Elba in 2014 for metabolomics (Supplementary Table 27 7). Samples were fixed in 1 mL cold methanol (HPLC-grade, Sigma-Aldrich) and stored at - 28 20°C until use. -

NST110: Advanced Toxicology Lecture 4: Phase I Metabolism
Absorption, Distribution, Metabolism and Excretion (ADME): NST110: Advanced Toxicology Lecture 4: Phase I Metabolism NST110, Toxicology Department of Nutritional Sciences and Toxicology University of California, Berkeley Biotransformation The elimination of xenobiotics often depends on their conversion to water-soluble chemicals through biotransformation, catalyzed by multiple enzymes primarily in the liver with contributions from other tissues. Biotransformation changes the properties of a xenobiotic usually from a lipophilic form (that favors absorption) to a hydrophilic form (favoring excretion in the urine or bile). The main evolutionary goal of biotransformation is to increase the rate of excretion of xenobiotics or drugs. Biotransformation can detoxify or bioactivate xenobiotics to more toxic forms that can cause tumorigenicity or other toxicity. Phase I and Phase II Biotransformation Reactions catalyzed by xenobiotic biotransforming enzymes are generally divided into two groups: Phase I and phase II. 1. Phase I reactions involve hydrolysis, reduction and oxidation, exposing or introducing a functional group (-OH, -NH2, -SH or –COOH) to increase reactivity and slightly increase hydrophilicity. O R1 - O S O sulfation O R2 OH Phase II Phase I R1 R2 R1 R2 - hydroxylation COO R1 O O glucuronidation OH R2 HO excretion OH O COO- HN H -NH2 R R1 R1 S N 1 Phase I Phase II COO O O oxidation glutathione R2 OH R2 R2 conjugation 2. Phase II reactions include glucuronidation, sulfation, acetylation, methylation, conjugation with glutathione, and conjugation with amino acids (glycine, taurine and glutamic acid) that strongly increase hydrophilicity. Phase I and II Biotransformation • With the exception of lipid storage sites and the MDR transporter system, organisms have little anatomical defense against lipid soluble toxins.