Methylation of the NT5E Gene Is Associated with Poor Prognostic Factors in Breast Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -

SOX9 Keeps Growth Plates and Articular Cartilage Healthy by Inhibiting Chondrocyte Dedifferentiation/ Osteoblastic Redifferentiation
SOX9 keeps growth plates and articular cartilage healthy by inhibiting chondrocyte dedifferentiation/ osteoblastic redifferentiation Abdul Haseeba,1, Ranjan Kca,1, Marco Angelozzia, Charles de Charleroya, Danielle Ruxa, Robert J. Towerb, Lutian Yaob, Renata Pellegrino da Silvac, Maurizio Pacificia, Ling Qinb, and Véronique Lefebvrea,2 aDivision of Orthopaedic Surgery, Children’s Hospital of Philadelphia, Philadelphia, PA 19104; bDepartment of Orthopaedic Surgery, University of Pennsylvania, Philadelphia, PA 19104; and cCenter for Applied Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104 Edited by Denis Duboule, University of Geneva, Geneva, Switzerland, and approved January 13, 2021 (received for review September 19, 2020) Cartilage is essential throughout vertebrate life. It starts develop- The skeleton is a model system to study cell fate and differ- ing in embryos when osteochondroprogenitor cells commit to entiation mechanisms. It arises developmentally from multi- chondrogenesis, activate a pancartilaginous program to form carti- potent mesenchymal cells, often called osteochondroprogenitors. laginous skeletal primordia, and also embrace a growth-plate pro- Guided by spatiotemporal cues, these cells commit to chondro- gram to drive skeletal growth or an articular program to build genesis or osteoblastogenesis to build cartilage or bone, respec- permanent joint cartilage. Various forms of cartilage malformation tively (5–7). Cartilage exists in several forms. Articular cartilage and degeneration diseases afflict humans, but underlying mecha- (AC) is a mostly resting tissue that protects opposing bone ends nisms are still incompletely understood and treatment options sub- in synovial joints throughout life, whereas growth plates (GPs) optimal. The transcription factor SOX9 is required for embryonic are transient, dynamic structures that drive skeletal growth while chondrogenesis, but its postnatal roles remain unclear, despite evi- being gradually replaced by bone (endochondral ossification). -

Transcriptomic and Proteomic Profiling Provides Insight Into
BASIC RESEARCH www.jasn.org Transcriptomic and Proteomic Profiling Provides Insight into Mesangial Cell Function in IgA Nephropathy † † ‡ Peidi Liu,* Emelie Lassén,* Viji Nair, Celine C. Berthier, Miyuki Suguro, Carina Sihlbom,§ † | † Matthias Kretzler, Christer Betsholtz, ¶ Börje Haraldsson,* Wenjun Ju, Kerstin Ebefors,* and Jenny Nyström* *Department of Physiology, Institute of Neuroscience and Physiology, §Proteomics Core Facility at University of Gothenburg, University of Gothenburg, Gothenburg, Sweden; †Division of Nephrology, Department of Internal Medicine and Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan; ‡Division of Molecular Medicine, Aichi Cancer Center Research Institute, Nagoya, Japan; |Department of Immunology, Genetics and Pathology, Uppsala University, Uppsala, Sweden; and ¶Integrated Cardio Metabolic Centre, Karolinska Institutet Novum, Huddinge, Sweden ABSTRACT IgA nephropathy (IgAN), the most common GN worldwide, is characterized by circulating galactose-deficient IgA (gd-IgA) that forms immune complexes. The immune complexes are deposited in the glomerular mesangium, leading to inflammation and loss of renal function, but the complete pathophysiology of the disease is not understood. Using an integrated global transcriptomic and proteomic profiling approach, we investigated the role of the mesangium in the onset and progression of IgAN. Global gene expression was investigated by microarray analysis of the glomerular compartment of renal biopsy specimens from patients with IgAN (n=19) and controls (n=22). Using curated glomerular cell type–specific genes from the published literature, we found differential expression of a much higher percentage of mesangial cell–positive standard genes than podocyte-positive standard genes in IgAN. Principal coordinate analysis of expression data revealed clear separation of patient and control samples on the basis of mesangial but not podocyte cell–positive standard genes. -

Cutting Edge: Adenosine A2a Receptor Signals Inhibit Germinal
Cutting Edge: Adenosine A2a Receptor Signals Inhibit Germinal Center T Follicular Helper Cell Differentiation during the Primary Response to Vaccination This information is current as of September 28, 2021. Shirdi E. Schmiel, Jessica A. Yang, Marc K. Jenkins and Daniel L. Mueller J Immunol published online 16 December 2016 http://www.jimmunol.org/content/early/2016/12/15/jimmun ol.1601686 Downloaded from Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on September 28, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published December 16, 2016, doi:10.4049/jimmunol.1601686 Th eJournal of Cutting Edge Immunology Cutting Edge: Adenosine A2a Receptor Signals Inhibit Germinal Center T Follicular Helper Cell Differentiation during the Primary Response to Vaccination Shirdi E. Schmiel,* Jessica A. Yang,† Marc K. Jenkins,† and Daniel L. Mueller* Adenosine A2a receptor (A2aR) signaling acts as a bar- asthma (10), autoimmunity (4, 5), and infection (11) also rier to autoimmunity by promoting anergy, inducing suggest that A2aR signaling can reduce Th1 and Th17 re- regulatory T cells, and inhibiting effector T cells. -

Directional Exosome Proteomes Reflect Polarity-Specific Functions in Retinal Pigmented Epithelium Monolayers
www.nature.com/scientificreports Correction: Author Correction OPEN Directional Exosome Proteomes Refect Polarity-Specifc Functions in Retinal Pigmented Epithelium Received: 9 February 2017 Accepted: 30 May 2017 Monolayers Published online: 07 July 2017 Mikael Klingeborn1, W. Michael Dismuke1, Nikolai P. Skiba1, Una Kelly1, W. Daniel Stamer1,2 & Catherine Bowes Rickman1,3 The retinal pigmented epithelium (RPE) forms the outer blood-retinal barrier in the eye and its polarity is responsible for directional secretion and uptake of proteins, lipoprotein particles and extracellular vesicles (EVs). Such a secretional division dictates directed interactions between the systemic circulation (basolateral) and the retina (apical). Our goal is to defne the polarized proteomes and physical characteristics of EVs released from the RPE. Primary cultures of porcine RPE cells were diferentiated into polarized RPE monolayers on permeable supports. EVs were isolated from media bathing either apical or basolateral RPE surfaces, and two subpopulations of small EVs including exosomes, and dense EVs, were purifed and processed for proteomic profling. In parallel, EV size distribution and concentration were determined. Using protein correlation profling mass spectrometry, a total of 631 proteins were identifed in exosome preparations, 299 of which were uniquely released apically, and 94 uniquely released basolaterally. Selected proteins were validated by Western blot. The proteomes of these exosome and dense EVs preparations suggest that epithelial polarity impacts directional release. These data serve as a foundation for comparative studies aimed at elucidating the role of exosomes in the molecular pathophysiology of retinal diseases and help identify potential therapeutic targets and biomarkers. Te retinal pigmented epithelium (RPE) is a cell monolayer that is situated between the photoreceptors and the systemic circulation of the choroid. -

Human CD Marker Chart Reviewed by HLDA1 Bdbiosciences.Com/Cdmarkers
BD Biosciences Human CD Marker Chart Reviewed by HLDA1 bdbiosciences.com/cdmarkers 23-12399-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD93 C1QR1,C1qRP, MXRA4, C1qR(P), Dj737e23.1, GR11 – – – – – + + – – + – CD220 Insulin receptor (INSR), IR Insulin, IGF-2 + + + + + + + + + Insulin-like growth factor 1 receptor (IGF1R), IGF-1R, type I IGF receptor (IGF-IR), CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD94 KLRD1, Kp43 HLA class I, NKG2-A, p39 + – + – – – – – – CD221 Insulin-like growth factor 1 (IGF-I), IGF-II, Insulin JTK13 + + + + + + + + + CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD178, FASLG, APO-1, FAS, TNFRSF6, CD95L, APT1LG1, APT1, FAS1, FASTM, CD95 CD178 (Fas ligand) + + + + + – – IGF-II, TGF-b latency-associated peptide (LAP), Proliferin, Prorenin, Plasminogen, ALPS1A, TNFSF6, FASL Cation-independent mannose-6-phosphate receptor (M6P-R, CIM6PR, CIMPR, CI- CD1d R3G1, R3 b-2-Microglobulin, MHC II CD222 Leukemia -

Lanthanum Breaks the Balance Between Osteogenesis and Adipogenesis of Mesenchymal Stem Cells Through Phosphorylation of Smad1/5/8
Electronic Supplementary Material (ESI) for RSC Advances. This journal is © The Royal Society of Chemistry 2015 Supporting Information Lanthanum Breaks the Balance between Osteogenesis and Adipogenesis of Mesenchymal Stem Cells through Phosphorylation of Smad1/5/8 Dandan Liu,ab Kun Ge,ac Jing Sun,c Shizhu Chen, ab Guang Jia ab and Jinchao Zhang * ab a College of Chemistry and Environmental Science, Chemical Biology Key Laboratory of Hebei Province, Hebei University, Baoding 071002, PR China b Key Laboratory of Medicinal Chemistry and Molecular Diagnosis of the Ministry of Education, Hebei University, Baoding 071002, PR China c Affiliated Hospital of Hebei University, Baoding 071000, PR China Table S1. The positions of 84 genes in supper array. Gene 1 2 3 4 5 6 7 8 9 10 11 12 A Abcb1a Alcam Anpep Anxa5 Bdnf Bglap1 Bmp2 Bmp4 Bmp6 Bmp7 Casp3 Cd44 B Col1a1 Csf2 Csf3 Ctnnb1 Egf Eng Erbb2 Fgf10 Fgf2 Fut1 Fut4 Fzd9 C Gdf15 Gdf5 Gdf6 Gdf7 Gtf3a Hat1 Hdac1 Hgf Hnf1a Icam1 Ifng Igf1 D Il10 Il1b Il6 Ins2 Itga6 Itgav Itgax Itgb1 Jag1 Kdr Kitl Lif E Mcam Mitf Mmp2 Nes Ngfr Notch1 Nt5e Nudt6 Kat2b Pdgfrb Pigs Pou5f1 F Pparg Prom1 Ptk2 Ptprc Rhoa Runx2 Slc17a5 Smad4 Smurf1 Smurf2 Sox2 Sox9 G Tbx5 Tert Tgfb1 Tgfb3 Thy1 Tnf Vcam1 Vegfa Vim Vwf Wnt3a Zfp42 H Gusb Hprt1 Hsp90ab1 Gapdh Actb MGDC RTC RTC RTC PPC PPC PPC Table S2. Genes involved in TGFβ/BMP signaling pathway are up-regulated in osteogenic differentiation of MSCs upon their interactions with 0.0001μM La. Functional GeneBank Gene Description Fold Symbol Change NM_007553 Bmp2 Bone morphogenetic -

Single-Cell RNA Landscape of Intratumoral Heterogeneity and Immunosuppressive Microenvironment in Advanced Osteosarcoma
ARTICLE https://doi.org/10.1038/s41467-020-20059-6 OPEN Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma Yan Zhou1,11, Dong Yang2,11, Qingcheng Yang2,11, Xiaobin Lv 3,11, Wentao Huang4,11, Zhenhua Zhou5, Yaling Wang 1, Zhichang Zhang2, Ting Yuan2, Xiaomin Ding1, Lina Tang 1, Jianjun Zhang1, Junyi Yin1, Yujing Huang1, Wenxi Yu1, Yonggang Wang1, Chenliang Zhou1, Yang Su1, Aina He1, Yuanjue Sun1, Zan Shen1, ✉ ✉ ✉ ✉ Binzhi Qian 6, Wei Meng7,8, Jia Fei9, Yang Yao1 , Xinghua Pan 7,8 , Peizhan Chen 10 & Haiyan Hu1 1234567890():,; Osteosarcoma is the most frequent primary bone tumor with poor prognosis. Through RNA- sequencing of 100,987 individual cells from 7 primary, 2 recurrent, and 2 lung metastatic osteosarcoma lesions, 11 major cell clusters are identified based on unbiased clustering of gene expression profiles and canonical markers. The transcriptomic properties, regulators and dynamics of osteosarcoma malignant cells together with their tumor microenvironment particularly stromal and immune cells are characterized. The transdifferentiation of malignant osteoblastic cells from malignant chondroblastic cells is revealed by analyses of inferred copy-number variation and trajectory. A proinflammatory FABP4+ macrophages infiltration is noticed in lung metastatic osteosarcoma lesions. Lower osteoclasts infiltration is observed in chondroblastic, recurrent and lung metastatic osteosarcoma lesions compared to primary osteoblastic osteosarcoma lesions. Importantly, TIGIT blockade enhances the cytotoxicity effects of the primary CD3+ T cells with high proportion of TIGIT+ cells against osteo- sarcoma. These results present a single-cell atlas, explore intratumor heterogeneity, and provide potential therapeutic targets for osteosarcoma. 1 Oncology Department of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, Shanghai 200233, China. -
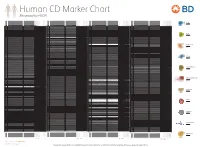
Reviewed by HLDA1
Human CD Marker Chart Reviewed by HLDA1 T Cell Key Markers CD3 CD4 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD8 CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD74 DHLAG, HLADG, Ia-g, li, invariant chain HLA-DR, CD44 + + + + + + CD158g KIR2DS5 + + CD248 TEM1, Endosialin, CD164L1, MGC119478, MGC119479 Collagen I/IV Fibronectin + ST6GAL1, MGC48859, SIAT1, ST6GALL, ST6N, ST6 b-Galactosamide a-2,6-sialyl- CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD75 CD22 CD158h KIR2DS1, p50.1 HLA-C + + CD249 APA, gp160, EAP, ENPEP + + tranferase, Sialo-masked lactosamine, Carbohydrate of a2,6 sialyltransferase + – – + – – + – – CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD75S a2,6 Sialylated lactosamine CD22 (proposed) + + – – + + – + + + CD158i KIR2DS4, p50.3 HLA-C + – + CD252 TNFSF4, -

CD System of Surface Molecules
THE CD SYSTEM OF LEUKOCYTE APPENDIX 4A SURFACE MOLECULES Monoclonal Antibodies to Human Cell Surface Antigens APPENDIX 4A Alice Beare,1 Hannes Stockinger,2 Heddy Zola,1 and Ian Nicholson1 1Women’s and Children’s Health Research Institute, Women’s and Children’s Hospital, Adelaide, Australia 2Institute of Immunology, University of Vienna, Vienna ABSTRACT Many of the leukocyte cell surface molecules are known by “CD” numbers. In this Appendix, a short introduction describes the history and the use of CD nomenclature and provides a few key references to enable access to the wider literature. This is followed by a table that lists all human molecules with approved CD names, tabulating alternative names, key structural features, cellular expression, major known functions, and usefulness of the molecules or antibodies against them in research or clinical applications. Curr. Protoc. Immunol. 80:A.4A.1-A.4A.73. C 2008 by John Wiley & Sons, Inc. Keywords: CD nomenclature r HLDA r HCDM r leukocyte marker r human leukocyte differentiation r antigens INTRODUCTION During the last 25 years, large numbers of monoclonal antibodies (MAbs) have been pro- duced that have facilitated the purification and functional characterization of a plethora of leukocyte surface molecules. The antibodies have been even more useful as markers for cell populations, allowing the counting, separation, and functional study of numer- ous subsets of cells of the immune system. A series of international workshops were instrumental in coordinating this development through multi-laboratory “blind” studies of thousands of antibodies. These HLDA (Human Leukocyte Differentiation Antigens) Workshops have, up until now, defined 500 different entities and assigned them cluster of differentiation (CD) designations. -

Integrin Activation Controls Regulatory T Cell–Mediated Peripheral Tolerance
Integrin Activation Controls Regulatory T Cell−Mediated Peripheral Tolerance Jane E. Klann, Stephanie H. Kim, Kelly A. Remedios, Zhaoren He, Patrick J. Metz, Justine Lopez, Tiffani Tysl, This information is current as Jocelyn G. Olvera, Jailal N. Ablack, Joseph M. Cantor, of September 24, 2021. Brigid S. Boland, Gene Yeo, Ye Zheng, Li-Fan Lu, Jack D. Bui, Mark H. Ginsberg, Brian G. Petrich and John T. Chang J Immunol 2018; 200:4012-4023; Prepublished online 27 April 2018; Downloaded from doi: 10.4049/jimmunol.1800112 http://www.jimmunol.org/content/200/12/4012 Supplementary http://www.jimmunol.org/content/suppl/2018/04/27/jimmunol.180011 http://www.jimmunol.org/ Material 2.DCSupplemental References This article cites 54 articles, 24 of which you can access for free at: http://www.jimmunol.org/content/200/12/4012.full#ref-list-1 Why The JI? Submit online. by guest on September 24, 2021 • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2018 by The American Association of Immunologists, Inc.