MIAMI UNIVERSITY the Graduate School Certificate for Approving The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Osr1 Maintains Renal Progenitors and Regulates Podocyte Development by Promoting Wnt2ba Through Antagonism of Hand2
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423845; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. osr1 maintains renal progenitors and regulates podocyte development by promoting wnt2ba through antagonism of hand2 Bridgette E. Drummond1, Brooke E. Chambers1, Hannah M. Wesselman1, Marisa N. Ulrich1, Gary F. Gerlach1, Paul T. Kroeger1, Ignaty Leshchiner2, Wolfram Goessling2, and Rebecca A. Wingert1* 1Department of Biological Sciences, Center for Stem Cells and Regenerative Medicine, Center for Zebrafish Research, University of Notre Dame, Notre Dame, 46556, USA 2Brigham and Women's Hospital, Genetics and Gastroenterology Division, Harvard Medical School, Harvard Stem Cell Institute, Boston, MA 02215, USA *Corresponding author Keywords: zebrafish, kidney, nephron, podocyte, segmentation, osr1, wnt2ba, hand2 Abbreviations: chronic kidney disease (CKD), N-ethyl-N-nitrosourea (ENU), days post fertilization (dpf), hours post fertilization (hpf), whole mount in situ hybridization (WISH), flourescent in situ hybridization (FISH), immunofluorescence (IF), immunocytochemistry (ICC), intermediate mesoderm (IM), congenital anomalies of the kidney and urinary tract (CAKUT), amino acid (aa), Zebrafish International Research Center (ZIRC), capped RNA (cRNA), somite stage (ss), wild-type (WT), LIM homeobox 1a (lhx1a), paired box 2a (pax2a), wilms tumor 1a (wt1a), wilms tumor 1b (wt1b), nephrosis 1, congenital Finnish type (nephrin) (nphs1), nephrosis 2, idiopathic, steroid-resistant (podocin)(nphs2), odd-skipped related transcription factor 1 (osr1), cadherin 17 (cdh17), odd-skipped related transcription factor 2 (osr2), wingless-type MMTV integration site family, member 2Ba (wnt2ba) and wingless-type MMTV integration site family, member 2Bb (wnt2bb), heart and neural crest derivatives expressed 2 (hand2), myosin light chain (myl7). -

The P63 Target HBP-1 Is Required for Keratinocyte Differentiation and Stratification
The p63 target HBP-1 is required for keratinocyte differentiation and stratification. Roberto Mantovani, Serena Borrelli, Eleonora Candi, Diletta Dolfini, Olì Maria Victoria Grober, Alessandro Weisz, Gennaro Melino, Alessandra Viganò, Bing Hu, Gian Paolo Dotto To cite this version: Roberto Mantovani, Serena Borrelli, Eleonora Candi, Diletta Dolfini, Olì Maria Victoria Grober, et al.. The p63 target HBP-1 is required for keratinocyte differentiation and stratification.. Cell Death and Differentiation, Nature Publishing Group, 2010, 10.1038/cdd.2010.59. hal-00542927 HAL Id: hal-00542927 https://hal.archives-ouvertes.fr/hal-00542927 Submitted on 4 Dec 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Running title: HBP1 in skin differentiation. The p63 target HBP-1 is required for skin differentiation and stratification. Serena Borrelli1, Eleonora Candi2, Bing Hu3, Diletta Dolfini1, Maria Ravo4, Olì Maria Victoria Grober4, Alessandro Weisz4,5, GianPaolo Dotto3, Gerry Melino2,6, Maria Alessandra Viganò1 and Roberto Mantovani1*. 1) Dipartimento di Scienze Biomolecolari e Biotecnologie. Università degli Studi di Milano. Via Celoria 26, 20133 Milano, Italy. 2) Biochemistry IDI-IRCCS laboratory, c/o University of Rome "Tor Vergata", Via Montpellier 1, 00133 Roma, Italy. -

Identification of SNP-Containing Regulatory Motifs in the Myelodysplastic Syndromes Model Using SNP Arrays Ad Gene Expression Arrays" (2013)
University of Central Florida STARS Faculty Bibliography 2010s Faculty Bibliography 1-1-2013 Identification of SNP-containing egulatr ory motifs in the myelodysplastic syndromes model using SNP arrays ad gene expression arrays Jing Fan Jennifer G. Dy Chung-Che Chang University of Central Florida Xiaoboo Zhou Find similar works at: https://stars.library.ucf.edu/facultybib2010 University of Central Florida Libraries http://library.ucf.edu This Article is brought to you for free and open access by the Faculty Bibliography at STARS. It has been accepted for inclusion in Faculty Bibliography 2010s by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Fan, Jing; Dy, Jennifer G.; Chang, Chung-Che; and Zhou, Xiaoboo, "Identification of SNP-containing regulatory motifs in the myelodysplastic syndromes model using SNP arrays ad gene expression arrays" (2013). Faculty Bibliography 2010s. 3960. https://stars.library.ucf.edu/facultybib2010/3960 Chinese Journal of Cancer Original Article Jing Fan 1, Jennifer G. Dy 1, Chung鄄Che Chang 2 and Xiaobo Zhou 3 Abstract Myelodysplastic syndromes have increased in frequency and incidence in the American population, but patient prognosis has not significantly improved over the last decade. Such improvements could be realized if biomarkers for accurate diagnosis and prognostic stratification were successfully identified. In this study, we propose a method that associates two state鄄of 鄄th e鄄ar t array technologies single nucleotide polymor鄄 要 phism (SNP) array and gene expression array with gene motifs considered transcription factor -binding 要 sites (TFBS). We are particularly interested in SNP鄄co ntaining motifs introduced by genetic variation and mutation as TFBS. -

An Order Estimation Based Approach to Identify Response Genes
AN ORDER ESTIMATION BASED APPROACH TO IDENTIFY RESPONSE GENES FOR MICRO ARRAY TIME COURSE DATA A Thesis Presented to The Faculty of Graduate Studies of The University of Guelph by ZHIHENG LU In partial fulfilment of requirements for the degree of Doctor of Philosophy September, 2008 © Zhiheng Lu, 2008 Library and Bibliotheque et 1*1 Archives Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington Ottawa ON K1A0N4 Ottawa ON K1A0N4 Canada Canada Your file Votre reference ISBN: 978-0-494-47605-5 Our file Notre reference ISBN: 978-0-494-47605-5 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library permettant a la Bibliotheque et Archives and Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par Plntemet, prefer, telecommunication or on the Internet, distribuer et vendre des theses partout dans loan, distribute and sell theses le monde, a des fins commerciales ou autres, worldwide, for commercial or non sur support microforme, papier, electronique commercial purposes, in microform, et/ou autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in et des droits moraux qui protege cette these. this thesis. Neither the thesis Ni la these ni des extraits substantiels de nor substantial extracts from it celle-ci ne doivent etre imprimes ou autrement may be printed or otherwise reproduits sans son autorisation. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

2017.08.28 Anne Barry-Reidy Thesis Final.Pdf
REGULATION OF BOVINE β-DEFENSIN EXPRESSION THIS THESIS IS SUBMITTED TO THE UNIVERSITY OF DUBLIN FOR THE DEGREE OF DOCTOR OF PHILOSOPHY 2017 ANNE BARRY-REIDY SCHOOL OF BIOCHEMISTRY & IMMUNOLOGY TRINITY COLLEGE DUBLIN SUPERVISORS: PROF. CLIONA O’FARRELLY & DR. KIERAN MEADE TABLE OF CONTENTS DECLARATION ................................................................................................................................. vii ACKNOWLEDGEMENTS ................................................................................................................... viii ABBREVIATIONS ................................................................................................................................ix LIST OF FIGURES............................................................................................................................. xiii LIST OF TABLES .............................................................................................................................. xvii ABSTRACT ........................................................................................................................................xix Chapter 1 Introduction ........................................................................................................ 1 1.1 Antimicrobial/Host-defence peptides ..................................................................... 1 1.2 Defensins................................................................................................................. 1 1.3 β-defensins ............................................................................................................. -
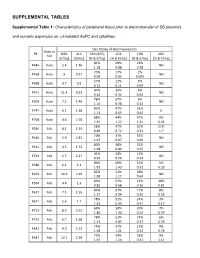
Supplemental Tables
SUPPLEMENTAL TABLES Supplemental Table 1: Characteristics of peripheral blood prior to electrotransfer of SB plasmids and numeric expansion on γ-irradiated AaPC and cytokines Day 0 (Day of electroporation) Auto or P# WBC ALC CD3 (ATC) CD4 CD8 ABC Allo [K/mL] [K/mL] [% & K/mL] [% & K/mL] [% & K/mL] [% & K/mL] 81% 60% 19% P446 Auto 5.4 1.46 ND 1.18 0.88 0.28 23% 17% 2% P458 Auto 9 0.27 ND 0.06 0.05 0.005 17% 12% 5% P468 Auto 4.7 0.9 ND 0.15 0.11 0.04 47% 35% 5% P471 Auto 11.4 0.93 ND 0.44 0.32 0.04 78% 67% 9% P509 Auto 7.2 1.46 ND 1.14 0.98 0.13 82% 47% 32% P747 Auto 6.1 1.38 0 1.13 0.65 0.44 88% 44% 47% 0% P708 Auto 4.6 2.78 2.45 1.22 1.31 0.05 58% 47% 20% 21% P396 Allo 8.1 1.54 0.89 0.72 0.31 1.7 70% 31% 32% P410 Allo 5.9 2.81 ND 1.97 0.87 0.90 80% 48% 32% P411 Allo 4.5 1.72 ND 1.38 0.83 0.55 41% 24% 15% P513 Allo 6.7 2.27 ND 0.93 0.54 0.34 86% 68% 15% 5% P580 Allo 6.1 2.1 1.81 1.43 0.32 0.29 82% 51% 28% P459 Allo 10.6 2.29 ND 1.88 1.17 0.64 64% 52% 12% 18% P564 Allo 4.4 1.3 0.83 0.68 0.16 0.81 82% 61% 17% 8% P617 Allo 7.1 1.55 1.27 0.94 0.26 0.58 78% 53% 24% 3% P671 Allo 5.4 1.7 1.33 0.90 0.41 0.17 69% 50% 20% 7% P723 Allo 8.6 2.61 1.80 1.30 0.52 0.59 78% 52% 22% 6% P732 Allo 6.7 1.68 1.31 0.87 0.37 0.39 74% 57% 15% 9% P641 Allo 9.0 2.13 1.58 1.21 0.32 0.78 73% 54% 18% 9% P647 Allo 12.1 2.29 1.67 1.24 0.41 1.11 75% 59% 11% P716 Allo 3.0 1.83 0 1.37 1.08 0.20 87% 73% 12% 1% P718 Allo 6.2 1.99 1.73 1.45 0.24 0.07 83% 69% 13% 6% P771 Allo 13.1 3.23 2.68 2.23 0.42 0.79 73% 42% 27% 6% P783 Allo 8.1 1.54 1.12 0.65 0.42 0.52 WBC = white blood -

Farnesol-Induced Apoptosis in Human Lung Carcinoma Cells Is Coupled to the Endoplasmic Reticulum Stress Response
Research Article Farnesol-Induced Apoptosis in Human Lung Carcinoma Cells Is Coupled to the Endoplasmic Reticulum Stress Response Joung Hyuck Joo,1 Grace Liao,1 Jennifer B. Collins,2 Sherry F. Grissom,2 and Anton M. Jetten1 1Cell Biology Section, LRB, and 2Microarray Group, Division of Intramural Research, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, North Carolina Abstract range of fruits and vegetables (9, 10). Each isoprenoid has been Farnesol (FOH) and other isoprenoid alcohols induce apopto- shown to inhibit proliferation and induce apoptosis in a number of sis in various carcinoma cells and inhibit tumorigenesis in neoplastic cell lines from different origins (4, 11–14). In addition, in vivo these isoprenoids have been reported to be effective in chemo- several models. However, the mechanisms by which in vivo they mediate their effects are not yet fully understood. In this prevention and chemotherapy in various cancer models study, we show that FOH is an effective inducer of apoptosis in (10, 12, 15, 16). FOH has been reported to exhibit chemopreventive several lung carcinoma cells, including H460. This induction is effects in colon and pancreas carcinogenesis in rats (9, 17) whereas associated with activation of several caspases and cleavage of phase I and II clinical trials have indicated therapeutic potential poly(ADP-ribose) polymerase (PARP). To obtain insight into for POH (16, 18). The mechanisms by which these isoprenoids induce these effects are not yet fully understood. Isoprenoids have the mechanism involved in FOH-induced apoptosis, we compared the gene expression profiles of FOH-treated and been reported to inhibit posttranslational protein prenylation (19) control H460 cells by microarray analysis. -

Temporal Chip-On-Chip of RNA-Polymerase-II to Detect Novel Gene Activation Events During Photoreceptor Maturation
Molecular Vision 2010; 16:252-271 <http://www.molvis.org/molvis/v16/a32> © 2010 Molecular Vision Received 12 July 2009 | Accepted 10 February 2010 | Published 17 February 2010 Temporal ChIP-on-Chip of RNA-Polymerase-II to detect novel gene activation events during photoreceptor maturation Padmaja Tummala, Raghuveer S. Mali, Eduardo Guzman, Xiao Zhang, Kenneth P. Mitton (The first two authors contributed equally to this work.) Eye Research Institute, Oakland University, Rochester, MI Purpose: During retinal development, post-mitotic neural progenitor cells must activate thousands of genes to complete synaptogenesis and terminal maturation. While many of these genes are known, others remain beyond the sensitivity of expression microarray analysis. Some of these elusive gene activation events can be detected by mapping changes in RNA polymerase-II (Pol-II) association around transcription start sites. Methods: High-resolution (35 bp) chromatin immunoprecipitation (ChIP)-on-chip was used to map changes in Pol-II binding surrounding 26,000 gene transcription start sites during photoreceptor maturation of the mouse neural retina, comparing postnatal age 25 (P25) to P2. Coverage was 10–12 kb per transcription start site, including 2.5 kb downstream. Pol-II-active regions were mapped to the mouse genomic DNA sequence by using computational methods (Tiling Analysis Software-TAS program), and the ratio of maximum Pol-II binding (P25/P2) was calculated for each gene. A validation set of 36 genes (3%), representing a full range of Pol-II signal ratios (P25/P2), were examined with quantitative ChIP assays for transcriptionally active Pol-II. Gene expression assays were also performed for 19 genes of the validation set, again on independent samples. -

Bioinformatics Studies Provide Insight Into Possible Target and Mechanisms of Action of Nobiletin Against Cancer Stem Cells
DOI:10.31557/APJCP.2020.21.3.611 Bioinformatics Studies Provide Insight into Possible Target and Mechanisms of Action of Nobiletin against Cancer Stem Cells RESEARCH ARTICLE Editorial Process: Submission:05/08/2019 Acceptance:03/06/2020 Bioinformatics Studies Provide Insight into Possible Target and Mechanisms of Action of Nobiletin against Cancer Stem Cells Adam Hermawan1*, Herwandhani Putri2 Abstract Objective: Nobiletin treatment on MDA-MB 231 cells reduces the expression of CXC chemokine receptor type 4 (CXCR4), which is highly expressed in cancer stem cell populations in tumor patients. However, the mechanisms of nobiletin in cancer stem cells (CSCs) remain elusive. This study was aimed to explore the potential target and mechanisms of nobiletin in cancer stem cells using bioinformatics approaches. Methods: Gene expression profiles by public COMPARE predicting the sensitivity of tumor cells to nobiletin. Functional annotations on gene lists are carried out with The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8, and WEB-based GEne SeT Analysis Toolkit (WebGestalt). The protein-protein interaction (PPI) network was analyzed by STRING-DB and visualized by Cytoscape. Results: Microarray analyses reveal many genes involved in protein binding, transcriptional and translational activity. Pathway enrichment analysis revealed breast cancer regulation of estrogen signaling and Wnt/ß-catenin by nobiletin. Moreover, three hub genes, i.e. ESR1, NCOA3, and RPS6KB1 and one significant module were filtered out and selected from the PPI network. Conclusion: Nobiletin might serve as a lead compound for the development of CSCs-targeted drugs by targeting estrogen and Wnt/ß-catenin signaling. Further studies are needed to explore the full therapeutic potential of nobiletin in cancer stem cells. -

UNIVERSITY of CALIFORNIA, IRVINE Combinatorial Regulation By
UNIVERSITY OF CALIFORNIA, IRVINE Combinatorial regulation by maternal transcription factors during activation of the endoderm gene regulatory network DISSERTATION submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biological Sciences by Kitt D. Paraiso Dissertation Committee: Professor Ken W.Y. Cho, Chair Associate Professor Olivier Cinquin Professor Thomas Schilling 2018 Chapter 4 © 2017 Elsevier Ltd. © 2018 Kitt D. Paraiso DEDICATION To the incredibly intelligent and talented people, who in one way or another, helped complete this thesis. ii TABLE OF CONTENTS Page LIST OF FIGURES vii LIST OF TABLES ix LIST OF ABBREVIATIONS X ACKNOWLEDGEMENTS xi CURRICULUM VITAE xii ABSTRACT OF THE DISSERTATION xiv CHAPTER 1: Maternal transcription factors during early endoderm formation in 1 Xenopus Transcription factors co-regulate in a cell type-specific manner 2 Otx1 is expressed in a variety of cell lineages 4 Maternal otx1 in the endodermal conteXt 5 Establishment of enhancers by maternal transcription factors 9 Uncovering the endodermal gene regulatory network 12 Zygotic genome activation and temporal control of gene eXpression 14 The role of maternal transcription factors in early development 18 References 19 CHAPTER 2: Assembly of maternal transcription factors initiates the emergence 26 of tissue-specific zygotic cis-regulatory regions Introduction 28 Identification of maternal vegetally-localized transcription factors 31 Vegt and OtX1 combinatorially regulate the endodermal 33 transcriptome iii -

P53 — 30 Yreviewsears On
FOCUS ON P53 — 30 YREVIEWSEARS ON p53 — a Jack of all trades but master of none Melissa R. Junttila and Gerard I. Evan Abstract | Cancers are rare because their evolution is actively restrained by a range of tumour suppressors. Of these p53 seems unusually crucial as either it or its attendant upstream or downstream pathways are inactivated in virtually all cancers. p53 is an evolutionarily ancient coordinator of metazoan stress responses. Its role in tumour suppression is likely to be a relatively recent adaptation, which is only necessary when large, long-lived organisms acquired the sufficient size and somatic regenerative capacity to necessitate specific mechanisms to reign in rogue proliferating cells. However, such evolutionary reappropriation of this venerable transcription factor entails compromises that restrict its efficacy as a tumour suppressor. Cnidarians–bilaterians Cancer is a genetic pathology that arises in the adult tissues questions are important for several reasons. Knowing Cinidarians comprise an animal of long-lived organisms, such as vertebrates, whose tis- how p53 discriminates between normal and tumour cells phylum of ~9,000 radially sues retain a substantial regenerative capacity throughout might point to attributes of cancer cells that qualitatively symmetrical, mostly marine life and, consequently, whose somatic cells accumulate distinguish them from normal somatic cells and could organisms. Most other animals are bilaterally symmetrical mutations. Occasionally, such mutations corrupt the be used as tumour-specific targets. Knowing what sig- and are classed as bilateria. regulatory mechanisms that suppress untoward somatic nals drive selection for loss of p53 function in cancers The cnidarians and bilaterians cell proliferation, survival and migration, resulting in the would help us understand what constrains and dictates last shared a common ancestor progressive outgrowth of a somatic clone.