Tissue Specific and Abiotic Stress Regulated Transcription of Histidine
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ectopic Expression of Nger Millet Calmodulin Confers Drought And
Ectopic expression of nger millet calmodulin confers drought and salinity tolerance in Arabidopsis thaliana Gautam Jamra University of Delhi - South Campus Aparna Agrawal G B Pant University of Agriculture and Technology: Govind Ballabh Pant University of Agriculture & Technology Nidhi Singh University of Delhi - South Campus Sibaji K. Sanyal University of Delhi - South Campus Anil Kumar G B Pant University of Agriculture and Technology: Govind Ballabh Pant University of Agriculture & Technology Girdhar Kumar Pandey ( [email protected] ) University of Delhi South Campus https://orcid.org/0000-0001-9180-0924 Research Article Keywords: Abiotic stress, calcium signalling, CaM, drought response, salinity response, overexpression Posted Date: April 13th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-383277/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at Plant Cell Reports on July 11th, 2021. See the published version at https://doi.org/10.1007/s00299-021-02743-z. Page 1/29 Abstract Drought and salinity are major environmental stresses which affect crop productivity and therefore are major hindrance in feeding growing population world-wide. Calcium (Ca2+) signalling plays a crucial role during the plant's response to these stress stimuli. Calmodulin (CaM), a crucial Ca2+sensor, is involved in transducing the signal downstream in various physiological, developmental and stress responses by modulating a plethora of target proteins. The role of CaM has been well established in the model plant Arabidopsis thaliana for regulating various developmental processes, stress signalling and ion transport. -

Forschungsbericht Für Das Jahr 2016
Forschungsbericht für das Jahr 2016 Fachbereich 13 - Biologie Impressum Herausgeber Westfälische Wilhelms-Universität Münster Prorektorin für Forschung Schlossplatz 2 48149 Münster E-Mail: [email protected] http://www.uni-muenster.de Bearbeitung und Layout Westfälische Wilhelms-Universität Münster Dezernat 6: Forschungsangelegenheiten Abteilung 6.4: Forschungsinformationen und Forschungsberichterstattung Domplatz 6-7 48143 Münster E-Mail: [email protected] http://www.uni-muenster.de/CRIS Abruf der Forschungsberichte http://www.uni-muenster.de/wwu/dokumentationen/forschungsberichte Münster, den 12.05.2017 Sehr geehrte Leserinnen und Leser, ein Forschungsbericht ist immer ein Spiegel geleisteter Arbeit und macht Eines deutlich: das starke Engagement der Forscherinnen und Forscher an der WWU. In ihren Forschungsberichten geben die Fachbereiche einen Überblick über die Forschungsaktivitäten des vergangenen Jahres 2016, über abgeschlossene Dissertations- und Habilitationsverfahren, über Publikationen, Auszeichnungen und Preise sowie nationale wie internationale Projekte, kurzum: über alles, was Forschung an der WWU auszeichnet. Dabei decken diese Aktivitäten ein breites Spektrum an Themen und Inhalten ab. So setzt sich das innovative Graduiertenkolleg EvoPAD („Evolutionary Processes in Adaptation and Disease”) aus geistes- und naturwissenschaftlicher Perspektive damit auseinander, wie und warum Krankheiten entstehen und welche Rolle evolutionäre Anpassungen dabei spielen. Während der SFB TRR 170 hingegen in höheren Sphären schwebt und unter anderem die Entwicklung der Erde und ihres Mondes ergründet, setzen sich gleich mehrere Projekte mit dem Thema der Digitalen Gesellschaft auseinander, beispielhaft auch aus genderspezifischer Sichtweise (z.B. EQUAL-IST oder Digital Me). Dies sind aber nur einige wenige Beispiele - die in diesem Bericht zusammengetragenen Daten zeigen die Vielfältigkeit aller Forschungsaktivitäten, die die WWU nicht nur bundesweit, sondern international sichtbar macht. -

Zone Wise List of NASI Fellows
The National Academy of Sciences, India (The Oldest Science Academy of India) Zone wise list of Fellows & Honorary Fellows (2021) 5, Lajpatrai Road, Prayagraj – 211002, UP, India 1 The list has been divided into six zones; and each zone is further having the list of scientists of Physical Sciences and Biological Sciences, separately. 2 The National Academy of Sciences, India 5, Lajpatrai Road, Prayagraj – 211002, UP, India Zone wise list of Fellows Zone 1 (Bihar, Jharkhand, Odisha, West Bengal, Meghalaya, Assam, Mizoram, Nagaland, Arunachal Pradesh, Tripura, Manipur and Sikkim) (Section A – Physical Sciences) ACHARYA, Damodar, Chairman, Advisory Board, SOA Deemed to be University, Khandagiri Squre, Bhubanesware - 751030; ACHARYYA, Subhrangsu Kanta, Emeritus Scientist (CSIR), 15, Dr. Sarat Banerjee Road, Kolkata - 700029; ADHIKARI, Satrajit, Sr. Professor of Theoretical Chemistry, School of Chemical Sciences, Indian Association for the Cultivation of Science, 2A & 2B Raja SC Mullick Road, Jadavpur, Kolkata - 700032; ADHIKARI, Sukumar Das, Formerly Professor I, HRI,Ald; Professor & Head, Department of Mathematics, Ramakrishna Mission Vivekananda University, Belur Math, Dist Howrah - 711202; BAISNAB, Abhoy Pada, Formerly Professor of Mathematics, Burdwan Univ.; K-3/6, Karunamayee Estate, Salt Lake, Sector II, Kolkata - 700091; BANDYOPADHYAY, Sanghamitra, Professor & Director, Indian Statistical Institute, 203, BT Road, Kolkata - 700108; BANERJEA, Debabrata, Formerly Sir Rashbehary Ghose Professor of Chemistry,CU; Flat A-4/6,Iswar Chandra Nibas 68/1, Bagmari Road, Kolkata - 700054; BANERJEE, Rabin, Emeritus Professor, SN Bose National Centre for Basic Sciences, Block - JD, Sector - III, Salt Lake, Kolkata - 700098; BANERJEE, Soumitro, Professor, Department of Physical Sciences, Indian Institute of Science Education & Research, Mohanpur Campus, WB 741246; BANERJI, Krishna Dulal, Formerly Professor & Head, Chemistry Department, Flat No.C-2,Ramoni Apartments, A/6, P.G. -

Department of Bio Technology
DEPARTMENT OF BIO TECHNOLOGY List of Ongoing Projects as on 08/07/2016 S.No. Title Project Investigator/Institute Date of Duration Total Cost Sanction (In Rs.) Agriculture Biotechnology – I 1 Programme Support for R&D in agricultural Prof. Anil Kumar Gupta 11/05/2012 5 Years 0 Month 27090000 Biotechnology-phase-II Programme at G.B. Pant G.B. Pant University of Agric. & Tech. University of Agriculture and Technology, Pant Pantnagar Pant Nagar, Uttarakhand- Nagar [BT/PR5031/AGR/2/850/2012] 263128 2 Program Support for Research and Development in Prof. R samiyappan 11/05/2012 5 Years 0 Month 31553004 Agricultural Biotechnology-Phase-II at Tamil Nadu Agricultural University TNAU,Coimbatore [BT/PR5095/AGR/2/847/2012] Coimbatore, Tamilnadu-641003 3 Characterization and molecular mapping of aphid Dr. Beant Singh 20/03/2015 3 Years 0 Month 5147600 (Rhopalosiphum maidis Fitch.) resistance in barley. Punjab Agricultural University [BT/PR9656/AGR/2/870/2013] Ludhiana, Punjab-141004 4 Emergence of Tobacco streak virus infecting Cotton Dr. Renukadevi P 20/04/2015 3 Years 0 Month 4970400 : Investigations on transmission, spread and Tamil Nadu Agricultural University symptom remission. Coimbatore, Tamilnadu-641003 [BT/PR11474/AGR/2/882/2014] 5 CHALLENGE PROGRAMME ON CHICKPEA Dr. Mukesh Jain 23/11/2015 5 Years 0 Month 78161600 FUNCTIONAL GENOMICS [BT/AGR/CG- National Institute of Plant Genome PhaseII/01/2014] Research New Delhi, Delhi-110067 Agriculture Biotechnology – II 6 Development of rice varieties for kerala with Dr. Jayalekshmy V G 02/08/2013 5 Years 0 Month 7240470 pyramided genes for Resistance to BLB by marker Kerala Agricultural University Thrissur, assisted selection [BT/PR13469/AGR/02/703/2010] Kerala-680656 7 Harnessing favorable QTL of wild and exotic Dr. -
Approaches to Generate Salinity Tolerant Cultivars of Rice
REVIEW published: 09 September 2015 doi: 10.3389/fpls.2015.00712 Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice Priyanka Das1,KamleshK.Nutan1, Sneh L. Singla-Pareek2 and Ashwani Pareek1* 1 Stress Physiology and Molecular Biology Laboratory, School of Life Sciences, Jawaharlal Nehru University, New Delhi, India, 2 Plant Molecular Biology Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, India Soil salinity is one of the main constraints affecting production of rice worldwide, by reducing growth, pollen viability as well as yield of the plant. Therefore, detailed understanding of the response of rice towards soil salinity at the physiological and molecular level is a prerequisite for its effective management. Various approaches have Edited by: been adopted by molecular biologists or breeders to understand the mechanism for Girdhar Kumar Pandey, University of Delhi, India salinity tolerance in plants and to develop salt tolerant rice cultivars. Genome wide Reviewed by: analysis using ‘omics-based’ tools followed by identification and functional validation Xinguang Zhu, of individual genes is becoming one of the popular approaches to tackle this task. On Chinese Academy of Sciences, China the other hand, mutation breeding and insertional mutagenesis has also been exploited Giridara Kumar Surabhi, Regional Plant Resource Centre, India to obtain salinity tolerant crop plants. This review looks into various responses at cellular *Correspondence: and whole plant level generated in rice plants toward salinity stress thus, evaluating Ashwani Pareek, the suitability of intervention of functional genomics to raise stress tolerant plants. We Stress Physiology and Molecular Biology Laboratory, School of Life have tried to highlight the usefulness of the contemporary ‘omics-based’ approaches Sciences, Jawaharlal Nehru such as genomics, proteomics, transcriptomics and phenomics towards dissecting out University, New Delhi 110067, India the salinity tolerance trait in rice. -

The Year Book 2020
THE YEAR BOOK 2020 INDIAN ACADEMY OF SCIENCES Bengaluru Postal Address: Indian Academy of Sciences Post Box No. 8005 C.V. Raman Avenue Sadashivanagar Post, Raman Research Institute Campus Bengaluru 560 080 India Telephone : +91-80-2266 1200, +91-80-2266 1203 Fax : +91-80-23616094 Email : [email protected], [email protected] Website : www.ias.ac.in © 2020 Indian Academy of Sciences Information in this Year Book is updated up to 31 January 2020. Editorial & Production Team: Nalini, B.R. Thirumalai, N. Vanitha, M. Published by: Executive Secretary, Indian Academy of Sciences Text formatted by WINTECS Typesetters, Bengaluru (Ph. +91-80-2332 7311) Printed by The Print Point, Bengaluru CONTENTS Page Section A: Indian Academy of Sciences Activities – a profile ................................................................. 2 Council for the period 2019–2021 ............................................ 6 Office Bearers ......................................................................... 7 Former Presidents ................................................................... 8 The Academy Trust ................................................................. 9 Section B: Professorships Raman Chair ........................................................................... 12 Jubilee Chair ........................................................................... 15 Janaki Ammal Chair ................................................................ 16 The Academy–Springer Nature Chair ...................................... 16 Section C: -
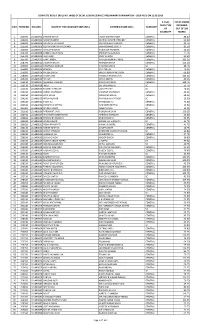
Marks Then Type Obtained S.No
COMPLETE RESULT (ROLL NO. WISE) OF DELHI JUDICIAL SERVICE PRELIMINARY EXAMINATION - 2019 HELD ON 22.09.2019 IF PwD, TOTAL MARKS THEN TYPE OBTAINED S.NO. FORM NO. ROLL NO. NAME OF THE CANDIDATE (MR./MS.) FATHER'S NAME (MR.) CATEGORY OF OUT OF 200 DISABILITY MARKS 1 260000 101490002 SHIVAM SINGH LAXMI SARAN SINGH GENERAL 88.00 2 266000 101490003 RAJESH KUMAR DEEPAK KUMAR CHOUBEY GENERAL 62.00 3 260200 101490004 KRITARTH KATHURIA MR. RAKESH KATHURIA GENERAL 43.75 4 253500 101490006 GUPTA RANI MANIKCHAND MANIKCHAND GUPTA GENERAL 81.50 5 264600 101490007 RAHUL PRASHAR RAJENDER PRASHAR GENERAL 44.25 6 253700 101490008 KANIKA AGGARWAL SHIVESH AGGARWAL GENERAL 77.75 7 254700 101490009 ISHA DHIR VIJAY KUMAR GENERAL 92.50 8 265700 101490010 AARTI JINDAL RAMESH KUMAR JINDAL GENERAL 103.25 9 258700 101490011 LAKSHAY BATRA MAHESH BATRA GENERAL 125.50 10 263800 101490012 VAISHNAVI SONKAR PAWAN KUMAR SC 88.75 11 255800 101490013 BARKHA RATTAN LAL GENERAL 74.25 12 256900 101490014 PIYUSH SINGH HRIDAY NARAYAN SINGH GENERAL 69.00 13 258900 101490015 AYUSH JAIN PRAVEEN KUMAR JAIN GENERAL 103.25 14 266010 101490016 HITESH DR K S NISHAL GENERAL 49.25 15 252110 101490017 ABHISHEK KAUSHIK RANVIR KAUSHIK GENERAL 108.75 16 253110 101490018 INDU SH. KRISHAN LAL SC 52.75 17 255110 101490019 YUGMITA PRATAP UDAI PRATAP SC 70.25 18 263210 101490020 ASHNA SACHDEVA PARVESH SACHDEVA GENERAL 46.25 19 262310 101490021 KRITI SINGH ARVINDER SINGH GENERAL 68.25 20 254310 101490022 NITISH KUMAR RAM KISHAN RATHOR SC 15.50 21 256410 101490023 SIDRA ALI MAHBOOB ALI GENERAL 73.00 -

Department of Plant Molecular Biology University of Delhi South Campus Estb
2018 Department of Plant Molecular Biology University of Delhi South Campus Estb. 1988 Cover Page About The mature wheat spikes represent the successful culmination of the Wheat Ge- The Department of Plant Molecular nome Sequencing Project in 2014, an international consortium project where Biology was established in 1988 under the DPMB contributed towards sequencing of the long arm of chromosome 2A. This Faculty of Interdisciplinary and Applied is the latest in a series of genome sequencing endeavors (starting from Rice Sciences to cater to the needs of students in 2005, followed by Tomato in 2012) to which our department has significantly in frontier areas of plant biology and to contributed. A graphical representation of a network on the right signifies current carry out research on Molecular Aspects and sustained efforts of all the faculty members of the DPMB towards under- of Plant Biology and Biotechnology. The standing the Gene regulatory networks, Protein-protein interactions and Signaling Department was enriched by merger networks that govern growth and development in plants and affect their ability to of the Unit for Plant Cell and Molecular withstand abiotic and biotic stress conditions. Biology in 1988 (originally established by the DST), and award of COSIST grant by the UGC (1990-1995). The Department has institutional as well as international been recognized for Special Assistance (2010 - 2013), and Professor Madan Mohan projects. The research has yielded about Programme (DRS Phase I to Phase III) (2013 - 2014) have served as Heads of the 750 publications and a few patents have Contacts by the UGC (2002-2018) to strengthen Department. -

The Year Book 2021
THE YEAR BOOK 2021 INDIAN ACADEMY OF SCIENCES Bengaluru Postal Address: Indian Academy of Sciences Post Box No. 8005 C.V. Raman Avenue Sadashivanagar Post, Raman Research Institute Campus Bengaluru 560 080 India Telephone : +91-80-2266 1200, +91-80-2266 1203 Fax : +91-80-2361 6094 Email : [email protected], [email protected] Website : www.ias.ac.in © 2021 Indian Academy of Sciences Information in this Year Book is updated up to 15 February 2021. Editorial & Production Team: Nalini, B.R. Thirumalai, N. Published by: Executive Secretary, Indian Academy of Sciences Text formatted by WINTECS Typesetters, Bengaluru (Mobile: 97310 01283) Printed by Ekakshara Printers, Bengaluru CONTENTS Page Section A: Indian Academy of Sciences Activities – a profile ................................................................. 2 Council for the period 2019–2021 ............................................ 6 Office Bearers ......................................................................... 7 Former Presidents ................................................................... 8 The Academy Trust ................................................................. 9 Section B: Professorships Raman Chair ........................................................................... 12 Jubilee Chair ........................................................................... 15 Janaki Ammal Chair ................................................................ 16 The Academy–Springer Nature Chair ...................................... 16 Section C: Fellowship -

Science & Engineering Research Board
Science & Engineering Research Board List of EMR Projects Sanctioned in the Year 2016-17 S. Total Discipline Sub Area Title PI Details No Cost 1. Chemical Inorganic & Investigation on the effect of ionic Dr. Harsh Kumar Manchanda, 3300000 Sciences Physical liquids on self assembly of various Department of Chemistry, Chemistry surfactants in Aqueous media Dr.B R Ambedkar National Institute of Technology, Jalandhar,Punjab, 144011. 2. Chemical Inorganic & Optical And Electrochemical Sensors Dr. Shweta Rana, 3025000 Sciences Physical based on Benzothiazole and their Department of Chemistry, Chemistry nanoparticles Panjab University, Chandigarh,160014. 3. Chemical Inorganic & A Concerted drive towards ambient Dr.Md. Motin Seikh, 3739560 Sciences Physical pressure synthesis and physical Department of Chemistry, Chemistry characterization of multiferroic Visva Bharati University, quadruple perovskites A'A3B4O12 Birbhum,West Bengal,731235. 4. Chemical Inorganic & Estimation of nanoparticles (Toxic Dr. Kalpana Kumari, 1900800 Sciences Physical metals) From River bed sediments of Department of Chemistry, Chemistry ganga and its tributries in vaishali and Birchand Patel Smarak College, muzaffarpur (Bihar) Vaishali,Bihar,844504. 5. Chemical Inorganic & Investigation on an Electrochemical Dr. Anudeep Kumar Narula, 3185600 Sciences Physical system comprising Guru Gobind Singh Indraprastha Chemistry polypyrrole/porphyrin/graphene-A University,New Delhi, potential approach towards dye 110006. sensitized solar cell and sensing 6. Chemical Inorganic & New Class of flexible solid-state Dr. Ramendra Sundar Dey, 2725050 Sciences Physical supercapacitor form nano-engineered Institute of Nano Science And Chemistry carbonaceous materisl Technology,Mohali, Punjab,160047. 7. Chemical Inorganic & Algrithmic implementation of beyond Prof. Satrajit Ahikari, 6167480 Sciences Physical born-Oppenheimer theories for Department of Physical Chemistry spectroscopic and scattering Chemistry, processes Indian Association for The Cultivation of Science, Kolkata,West Bengal,700032. -

For Ramie (Boehmeria Nivea L. Gaud)
Development and Characterization of 1,827 Expressed Sequence Tag-Derived Simple Sequence Repeat Markers for Ramie (Boehmeria nivea L. Gaud) Touming Liu1, Siyuan Zhu1, Lili Fu1, Qingming Tang1, Yongting Yu1, Ping Chen1, Mingbao Luan1, Changbiao Wang2, Shouwei Tang1* 1 Institute of Bast Fiber Crops and Center of Southern Economic Crops, Chinese Academy of Agricultural Sciences, Changsha, China, 2 Biotechnology Research Center, Shanxi Academy of Agricultural Sciences, Taiyuan, China Abstract Ramie (Boehmeria nivea L. Gaud) is one of the most important natural fiber crops, and improvement of fiber yield and quality is the main goal in efforts to breed superior cultivars. However, efforts aimed at enhancing the understanding of ramie genetics and developing more effective breeding strategies have been hampered by the shortage of simple sequence repeat (SSR) markers. In our previous study, we had assembled de novo 43,990 expressed sequence tags (ESTs). In the present study, we searched these previously assembled ESTs for SSRs and identified 1,685 ESTs (3.83%) containing 1,878 SSRs. Next, we designed 1,827 primer pairs complementary to regions flanking these SSRs, and these regions were designated as SSR markers. Among these markers, dinucleotide and trinucleotide repeat motifs were the most abundant types (36.4% and 36.3%, respectively), whereas tetranucleotide, pentanucleotide, and hexanucleotide motifs represented ,10% of the markers. The motif AG/CT was the most abundant, accounting for 28.74% of the markers. One hundred EST- SSR markers (97 SSRs located in genes encoding transcription factors and 3 SSRs in genes encoding cellulose synthases) were amplified using polymerase chain reaction for detecting 24 ramie varieties. -

Protein Phosphatase 2A in the Regulatory Network Underlying Biotic Stress Resistance in Plants
fpls-07-00812 June 8, 2016 Time: 13:25 # 1 CORE Metadata, citation and similar papers at core.ac.uk Provided by Frontiers - Publisher Connector REVIEW published: 10 June 2016 doi: 10.3389/fpls.2016.00812 Protein Phosphatase 2A in the Regulatory Network Underlying Biotic Stress Resistance in Plants Guido Durian1, Moona Rahikainen1, Sara Alegre1, Mikael Brosché2 and Saijaliisa Kangasjärvi1* 1 Department of Biochemistry, Molecular Plant Biology, University of Turku, Turku, Finland, 2 Division of Plant Biology, Department of Biosciences, Viikki Plant Science Centre, University of Helsinki, Helsinki, Finland Biotic stress factors pose a major threat to plant health and can significantly deteriorate plant productivity by impairing the physiological functions of the plant. To combat the wide range of pathogens and insect herbivores, plants deploy converging signaling pathways, where counteracting activities of protein kinases and phosphatases form a basic mechanism for determining appropriate defensive measures. Recent studies have identified Protein Phosphatase 2A (PP2A) as a crucial component that controls pathogenesis responses in various plant species. Genetic, proteomic and metabolomic approaches have underscored the versatile nature of PP2A, which contributes to the regulation of receptor signaling, organellar signaling, gene expression, metabolic pathways, and cell death, all of which essentially impact plant immunity. Associated with Edited by: this, various PP2A subunits mediate post-translational regulation of metabolic enzymes Olivier Lamotte, UMR Agroécologie, France and signaling components. Here we provide an overview of protein kinase/phosphatase Reviewed by: functions in plant immunity signaling, and position the multifaceted functions of PP2A Alison DeLong, in the tightly inter-connected regulatory network that controls the perception, signaling Brown University, USA and responding to biotic stress agents in plants.