Genome-Wide Analysis of the Diversity and Ancestry of Korean Dogs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NEW DOG BREEDS RECOGNIZED by the FCI THAI BANGKAEW DOG Text and Illustrations by RIA HÖRTER Photos Courtesy of Mr
HISTORY The FCI (Fédération Cynologique Interna- lation. A Buddhist abbot played an important role in tionale), the World Canine Organization, includes 86 the history of the Thai Bangkaew Dog. member countries and contract partners (one mem- There are 76 individual living languages in Thai- ber per country). Each issues its own pedigrees and land. The official language is Thai, a so-called tone trains its own judges. The FCI ensures that the pedi- language. In Thai, every syllable is pronounced in grees and judges are mutually recognized by all FCI one of five tones: low, mid, high, falling or rising. members. The Thai written language is essentially alphabetic, Recognition of a breed by the FCI means that in but notoriously difficult to read. almost every European country, that breed can be awarded FCI championship prizes. One of the newly ANCIENT RELATION recognized breeds is the Thai Bangkaew Dog. From the 10th century onward, Tai people (of which the Thai are a subgroup) travelled from south Thailand’s Thai Bangkaew Dog is classified by China to settle in what is now Thailand, where they NEW DOG BREEDS RECOGNIZED BY THE FCI THAI BANGKAEW DOG text and illustrations by RIA HÖRTER Photos Courtesy Of Mr. Jetsada Sangjan the FCI in Group 5, Spitz and primitive type; Section merged with local tribes, such as the Khmers and 5, Asian Spitzes and related breeds. The official stan- Mons. No doubt their dogs accompanied them in the dard was published in April, 2013. migration. From the 13th century, the Thai culture was dominant in Thailand. -

Dog Breeds of the World
Dog Breeds of the World Get your own copy of this book Visit: www.plexidors.com Call: 800-283-8045 Written by: Maria Sadowski PlexiDor Performance Pet Doors 4523 30th St West #E502 Bradenton, FL 34207 http://www.plexidors.com Dog Breeds of the World is written by Maria Sadowski Copyright @2015 by PlexiDor Performance Pet Doors Published in the United States of America August 2015 All rights reserved. No portion of this book may be reproduced or transmitted in any form or by any electronic or mechanical means, including photocopying, recording, or by any information retrieval and storage system without permission from PlexiDor Performance Pet Doors. Stock images from canstockphoto.com, istockphoto.com, and dreamstime.com Dog Breeds of the World It isn’t possible to put an exact number on the Does breed matter? dog breeds of the world, because many varieties can be recognized by one breed registration The breed matters to a certain extent. Many group but not by another. The World Canine people believe that dog breeds mostly have an Organization is the largest internationally impact on the outside of the dog, but through the accepted registry of dog breeds, and they have ages breeds have been created based on wanted more than 340 breeds. behaviors such as hunting and herding. Dog breeds aren’t scientifical classifications; they’re It is important to pick a dog that fits the family’s groupings based on similar characteristics of lifestyle. If you want a dog with a special look but appearance and behavior. Some breeds have the breed characterics seem difficult to handle you existed for thousands of years, and others are fairly might want to look for a mixed breed dog. -

THE NORDIC SHOW Hosted by the Finnish Spitz Club for Spitz Breeds SCHEDULE of 152 Class Unbenched
THE NORDIC SHOW Hosted by The Finnish Spitz Club for Spitz Breeds SCHEDULE of 152 Class Unbenched Sponsored by Sponsored by OPEN SHOW (Not judged on the Group System) (Held under Kennel Club Limited Rules and Regulations) at THE SPORTS CONNEXION Leamington Road, Ryton on Dunsmore, nr Coventry CV8 3FL on SATURDAY, 27th NOVEMBER 2021 Show opens: 8.30 am Judging: Junior Handling at 9.00 am; Breed at 9.30 am Show closes half an hour after completion of judging JURISDICTION AND RESPONSIBILITIES: The Officers and Committee members of the society holding the licence are deemed responsible for organising and conducting the show safely and in accordance with the Rules and Regulations of the Kennel Club and agree to abide by and adopt any decision of the Board or any authority to whom the Board may delegate its powers subject to the conditions of Regulation F17. In so doing those appointed as officers and committee members accept that they are jointly and severally responsible for the organisation of the show and that this is a binding undertaking (vide Kennel Club General Show Regulations F4 and F5). Hon. Veterinary Surgeon (on call): Medivet The Vets 1 Guy Street, Leamington Spa, Warks CV32 4RX. Tel: 01926 423161 Show Manager/Covid Safety Officer: Mr Stuart Byrne All Judges at this show agree to abide by the following statement: “In assessing dogs, judges must penalise any features or exaggerations which they consider would be detrimental to the soundness, health and well being of the dog.” Postal entries close: MONDAY, 1st NOVEMBER 2021 (Postmark) On-line entries can be made up until midnight on Sunday, 7th November 2021 at www.fossedata.co.uk Postal entries and fees to be sent to the Hon. -

MSK Dog EN Letterus.Indd
Most Common Breeds size: S = small; M = medium; L = large; G = giant lifespan: A = approx. 9 years; B = approx. 11 years; C = approx. 15 years hound group gundog group breed size lifespan breed size lifespan Afghan Hound L B Bracco Italiano L B Azawakh L B Brittany M B Basenji M B English Setter L B Basset Bleu de Gascogne M B German Long-Haired Pointer L B Basset Fauve de Bretagne M B German Short-Haired Pointer L B Basset Griffon Vendeen (Grand) M B German Wire-Haired Pointer L B Basset Griffon Vendeen (Petit) M B Gordon Setter L B Basset Hound M B Hungarian Vizsla L B Bavarian Mountain Hound M B Hungarian Wire-Haired Vizsla L B Beagle M B Irish Red and White Setter L B Bloodhound L A Irish Setter L B Borzoi L B Italian Spinone L B Cirneco dell'Etna M B Kooikerhondje M B Dachshund (Long-Haired) M B Korthals Griffon L B Dachshund (Miniature Long-Haired) S C Lagotto Romagnolo M B Dachshund (Smooth-Haired) M B Large Munsterlander L B Dachshund (Miniature Smooth-Haired) S C Pointer L B Dachshund (Wire-Haired) M B Retriever (Chesapeake Bay) L B Dachshund (Miniature Wire-Haired) S C Retriever (Curly-Coated) L B Deerhound L B Retriever (Flat-Coated) L B Finnish Spitz M B Retriever (Golden) L B Foxhound L B Retriever (Labrador) L B Grand Bleu de Gascogne L B Retriever (Nova Scotia Duck-Tolling) M B Greyhound L B Slovakian Rough-Haired Pointer L B Hamiltonstovare L B Small Munsterlander M B Ibizan Hound L B Spaniel (American Cocker) M B Irish Wolfhound G A Spaniel (Clumber) L B Norwegian Elkhound L B Spaniel (Cocker) M B Otterhound L B Spaniel -

New Jersey Animal Guidelines
New Jersey Animal Guidelines: Any of the following animals owned, kept by, in the care, custody or control of any occupants of the home are ineligible: 1. Any animal deemed dangerous, vicious or potentially dangerous under state statute. 2. Any exotic animal, wild or zoo animals (including but not limited to reptiles, primates, exotic cats and fowl). 3. Any of the following dogs: • Akita Inu • German Shepherd • Alaskan Malamute • Giant Schnauzer • American Bull Dog • Great Dane • American Eskimo Dog (member of the • Gull Dong (aka Pakistani Bull Dog) Spitz Family) • American Staffordshire Terrier • Gull terrier • American Put Bull Terrier • Husky or Siberian Husky • Beauceron • Japanese Tosa/Tosa Inu/Tosa Ken • Boerboel • Korean Jindo • Bull Mastiff/American Bandogge/Bully • Perro de Presa Canario Kutta (any other Mastiff breed) • Cane Corso • Perro de Presa Mallorquin • Caucasian Ovcharka (Mountain Dog) • “Pit Bull” • Chow Chow • Rottweiler • Doberman Pinsher (other than a • Rhodesian Ridgeback miniature Doberman • Dogo Argentino • Staffordshire Bull Terrier • English Bull Terrier • Thai Ridgeback • Fila Brasileiro (aka Brazilian Mastiff) • Wolf or Wolf Hybrid Or any mixed breed dog containing any of the aforementioned breeds. 4. A dog that has been trained as and/or used as a guard dog or attack dog. 5. A dog that has been trained or used by the military or police for enforcing public order by chasing and holding suspects by the threat of being released, either by direct apprehension or a method known as “Bark and Hold”. 6. A dog belonging to a breed that was historically bred for fighting. 7. A dog that has bitten anyone or has exhibited aggressive behavior towards people. -

Dog Breeds Pack 1 Professional Vector Graphics Page 1
DOG BREEDS PACK 1 PROFESSIONAL VECTOR GRAPHICS PAGE 1 Affenpinscher Afghan Hound Aidi Airedale Terrier Akbash Akita Inu Alano Español Alaskan Klee Kai Alaskan Malamute Alpine Dachsbracke American American American American Akita American Bulldog Cocker Spaniel Eskimo Dog Foxhound American American Mastiff American Pit American American Hairless Terrier Bull Terrier Staffordshire Terrier Water Spaniel Anatolian Anglo-Français Appenzeller Shepherd Dog de Petite Vénerie Sennenhund Ariege Pointer Ariegeois COPYRIGHT (c) 2013 FOLIEN.DS. ALL RIGHTS RESERVED. WWW.VECTORART.AT DOG BREEDS PACK 1 PROFESSIONAL VECTOR GRAPHICS PAGE 2 Armant Armenian Artois Hound Australian Australian Kelpie Gampr dog Cattle Dog Australian Australian Australian Stumpy Australian Terrier Austrian Black Shepherd Silky Terrier Tail Cattle Dog and Tan Hound Austrian Pinscher Azawakh Bakharwal Dog Barbet Basenji Basque Basset Artésien Basset Bleu Basset Fauve Basset Griffon Shepherd Dog Normand de Gascogne de Bretagne Vendeen, Petit Basset Griffon Bavarian Mountain Vendéen, Grand Basset Hound Hound Beagle Beagle-Harrier COPYRIGHT (c) 2013 FOLIEN.DS. ALL RIGHTS RESERVED. WWW.VECTORART.AT DOG BREEDS PACK 2 PROFESSIONAL VECTOR GRAPHICS PAGE 3 Belgian Shepherd Belgian Shepherd Bearded Collie Beauceron Bedlington Terrier (Tervuren) Dog (Groenendael) Belgian Shepherd Belgian Shepherd Bergamasco Dog (Laekenois) Dog (Malinois) Shepherd Berger Blanc Suisse Berger Picard Bernese Mountain Black and Berner Laufhund Dog Bichon Frisé Billy Tan Coonhound Black and Tan Black Norwegian -

A Review of the Jindo, Korean Native Doga -Review
381 A Review of the Jindo, Korean Native Doga -Review - C. G. Lee*, J. I. Lee, C. Y. Lee and S. S. Sun1* College of Veterinary Medicine, Chonnam National University, Kwangju 500-757, Korea ABSTRACT : The Jindo is a Korean native dog, well-known for its hunting and guarding abilities. When he gives his devotion to one individ니 al, he gives it whole-heartedly. He is not tempted easily and impetuous. The breed was not developed, b니 t the dog retained their original qualities -loyal, alert, fearless, obedient, watchful, intelligent, energetic- to survive in the harsh environment of the Jindo island. The dog had been spread over the entire Korean peninsula from the time unknown, and the ones in the Jindo island, isolated until lately, survived and maintained their original characteristics. They are now spread over the entire Jindo County consisted of many islands, whence the breed natne came. The Jindo comes in a variety of colors and color combinations, with the fawn and white colorings predominant. The dog is one of the Korean natural monuments, protected by law since early 1960s. The Jindo gained official approval by the Federation Cynologique Internationale as a hunting dog. Apart from the basic housetraining, the dog rarely gets training. Many people have attempted to preserve its pure bloodlines and original qualities. Today, there are a total of 10,356 Jindoes being raised over the entire Jindo County, and many more are kept elsewhere. A research into genetic characteristics of the Jindo is now goin은 on, using the technique of isozyme electrophoresis. The Jindo Dog Breeding Management Center has been reinforced lately, and in addition to their routines, the Center is to work on the breeding of the Jindo. -
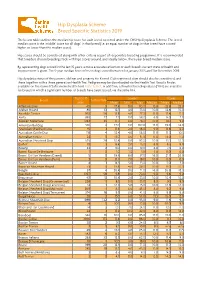
Hip Dysplasia Scheme Breed Specific Statistics 2019
Hip Dysplasia Scheme Breed Specific Statistics 2019 The below table outlines the median hip score for each breed screened under the CHS Hip Dysplasia Scheme. The breed median score is the ‘middle’ score for all dogs’ in that breed (i.e. an equal number of dogs in that breed have scored higher or lower than the median score). Hip scores should be considered along with other criteria as part of responsible breeding programme. It is recommended that breeders choose breeding stock with hips scores around, and ideally below, the 5-year breed median score. By representing dogs scored in the last 15 years, a more accurate reflection of each breed’s current state of health and improvement is given. The 5-year median here refers to dogs scored between 1st January 2015 and 31st December 2019. Hip dysplasia status of the parents, siblings and progeny for Kennel Club registered dogs should also be considered, and these together with a three generation Health Test Pedigree may be downloaded via the Health Test Results Finder, available on the Kennel Club's online health tool Mate Select. In addition, estimated breeding values (EBVs) are available for breeds in which a significant number of breeds have been scored, via the same link. Tested 15 15 years 5 years Breed Tested 2019 years Mean Min Max Median Mean Median Affenpinscher 40 0 17.9 8.0 90.0 13.0 23.8 23.0 Afghan Hound 85 33 12.3 4.0 73.0 10.0 12.6 10.0 Airedale Terrier 910 58 13.9 4.0 77.0 11.0 13.8 11.0 Akita 883 27 7.7 0.0 58.0 6.0 8.0 7.0 Alaskan Malamute 1242 25 11.7 0.0 78.0 10.0 10.1 9.0 -

Mtdna D-Loop의 염기서열에 의한 제주견과 우리나라 재래견 및 외국견 품종과의 유연관계 Phylogenetic
Journal of Animal Science and Technology 53(4) 303~310, 2011 http://dx.doi.org/10.5187/JAST.2011.53.4.303 mtDNA D-loop의 염기서열에 의한 제주견과 우리나라 재래견 및 외국견 품종과의 유연관계 김미경1․김남영2․이성수2․김규일1․양영훈1* 1제주대학교 생명공학부, 2농촌진흥청 난지축산시험장 Phylogenetic Relationships of Jeju Dogs to Other Domestic and Foreign Dog Breeds Determined by Using mtDNA D-loop Sequences Mi Gyoung Kim1, Nam Young Kim2, Sung Soo Lee2, Ky IL Kim1 and Young Hoon Yang1* 1Faculty of Biotechnology, Jeju National University, Jejudaekakno 66, Jeju-si, Jeju 690-756, Korea, 2National Institute of Animal Science RDA, O-deung Dong 175-6, Jeju 690-150, Korea ABSTRACT Phylogenetic relationships of Jeju dogs to other domestic and foreign dog breeds were assessed using mtDNA D-loop sequences. Neighbor-joining trees were constructed using complete sequences (970 bp excluding the tandem repeat region) determined for five Cheju, four Jindo, four Sapsaree, five Pungsan, two of each East and West Laika dogs (Canis familiaris), two gray wolves (Canis lupus) and two coyotes (Canis latrans) and also published complete sequences for dogs. Coyote sequences were used as outgroups. In addition, a total of 214 haplotypes of 598bp D-loop sequences from 30 dog breeds were collected from GenBank and used to investigate genetic structure of population. In the analyses of full D-loop sequence variation and the phylogenetic trees constructed by neighbor-joining method, neither haplotypes nor clades specific for any domestic dog breeds were observed. The inter-species sequence variation (4.51%) between domestic dogs and wolves was much higher than the intra-species sequence variation within domestic dogs (1.63%) and wolves (3.64%). -

Specified Records Related to the Number and Breed of Aggressive Dogs in Vancouver
~YOF CITY CLERK'S DEPARTMENT VANCOUVER Information, Admi nist ration and Election Services File No. 04-1000-20-2016-351 October 26, 201 6 Re: Request for Access to Records under the Freedom of Information and Protection of Privacy Act (the "Act") I am writing in response to your request of September 15, 2016 under the Freedom of Information and Protection of Privacy Act, (the Act), for: The following records as of August 31, 2016: 1) Number of dogs registered as dangerous/ vicious/ aggressive in the city of Vancouver; 2) The breed of each dog registered as such; 3) The number of repeat owners on the list (has an owner had more than one dog registered on the list - if so, how many). All responsive records the City is able to provide are at tached. This data has been compiled by reviewing each dog license file and manually recording the dog breed for files where the dog was flagged as aggressive. The search encompassed all dogs residing in Vancouver in 2016 which are currently licensed or tagged as requiring renewal. Please note: Animal Control is not able to compile a report on dog owners who have had more than one aggressive dog registered to t hem historically. The current animal control database has efficiency and data reporting limitations, it is gradually being phased out. Please refer to the letter sent to you on October 21, 201 6 for further det ai l. If you request a review, please provide the Commissioner's office with: 1) the request number assigned to your request (#04-1000-20-201 6-351); 2) a copy of this letter; 3) a copy of your original request for information sent to the City of Vancouver; and 4) detailed reasons or grounds on which you are seeking the review. -

Rescue Partners
COUNTY OF SAN DIEGO DEPARTMENT OF ANIMAL SERVICES RESCUE PARTNER BREED LISTING OCTOBER 2019 Table of Contents BIRDS ................................................................................................................................................... 2 CATS .................................................................................................................................................... 3 DOGS ................................................................................................................................................... 5 EQUINES ............................................................................................................................................ 16 FARM ................................................................................................................................................. 17 FISH ................................................................................................................................................... 18 RABBITS & SMALL ANIMALS ................................................................................................................ 19 REPTILES ............................................................................................................................................ 20 RESCUE ORGANIZATIONS (Alphabetical Listing) ............................................................................... 21 A ....................................................................................................................................................... -

Canis-Adn-Mix Razas Caninas
CANIS-ADN-MIX RAZAS CANINAS Affenpinscher Bohemian Shepherd Afghan Hound Bolognese Airedale Terrier Border Collie Akita Border Terrier Alaskan Klee Kai Borzoi Alaskan Malamute Boston Terrier Alpine Dachsbracke Bouvier Des Flanders American English Coonhound Boxer American Eskimo Dog Boykin Spaniel American Foxhound Bracco Italiano American Hairless Rat Terrier Braque du Bourbonnais American Indian Dog Brazilian Terrier American Staffordshire Terrier Briard American Water Spaniel Brussels Griffon Anatolian Shepherd Dog Bulgarian Shepherd Appenzell Cattle Dog Bull Arab Argentine Dogo Bull Terrier (Miniature) Australian Cattle Dog Bull Terrier (Standard) Australian Kelpie Bulldog (American) Australian Labradoodle Bulldog (Standard) Australian Shepherd Bullmastiff Australian Stumpy Tail Cattle Dog Cairn Terrier Australian Terrier Canaan Dog Azawakh Canadian Eskimo Dog Barbet Cane Corso Basenji Cardigan Welsh Corgi Basset Bleu de Gascogne Catahoula Leopard Dog Basset Fauve de Bretagne Catalan Sheepdog Basset Griffon Vendeen (Grand) Caucasian Shepherd Dog Basset Griffon Vendeen (Petit) Cavalier King Charles Spaniel Basset Hound Central Asian Ovcharka Bavarian Mountain Hound Cesky Terrier Beagle Chesapeake Bay Retriever Bearded Collie Chihuahua Beauceron Chinese Crested Bedlington Terrier Chinese Shar-Pei Belgian Malinois Chinook Belgian Mastiff Chow Chow Belgian Sheepdog Cirneco del Etna Belgian Tervuren Clumber Spaniel Bergamasco Cocker Spaniel Berger Picard Collie Bernese Mountain Dog Continental Toy Spaniel (phalene and papillon) Bichon