De Duve's Peroxisomes and Meyer's Hydrogenases in The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chloroplast Transit Peptides: Structure, Function and Evolution
reviews Chloroplast transit Although the first demonstration of precursor trans- port into chloroplasts was shown over two decades peptides: structure, ago3,4, only now is this area of cell biology becom- ing well understood. Many excellent reviews have been published recently on the evolution of plas- function and tids5, the evolution of organelle genomes6, the mechanism of gene transfer from organelles to the nucleus7 and the mechanism of protein import into evolution chloroplasts8,9. Proteins destined to plastids and other organ- elles share in common the requirement for ‘new’ Barry D. Bruce sequence information to facilitate their correct trafficking within the cell. Although in most cases this information resides in a cleavable, N-terminal sequence often collectively referred to as signal It is thought that two to three thousand different proteins are sequence, the different organelle-targeting se- targeted to the chloroplast, and the ‘transit peptides’ that act as quences have distinct properties and names: ‘signal peptides’ for the endoplasmic reticulum, chloroplast targeting sequences are probably the largest class of ‘presequences’ for the mitochondria and ‘transit peptides’ for chloroplasts and other plastids. This targeting sequences in plants. At a primary structural level, transit review focuses on recent progress in dissecting peptide sequences are highly divergent in length, composition and the role of the stromal-targeting domain of chloro- plast transit peptides. I will consider briefly the organization. An emerging concept suggests that transit peptides multitude of distinct functions that transit peptides contain multiple domains that provide either distinct or overlapping perform, provide an update on the limited struc- tural information of a number of transit peptides functions. -

Centrosome-Nuclear Envelope Tethering and Microtubule Motor
bioRxiv preprint doi: https://doi.org/10.1101/442368; this version posted October 12, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Centrosome-nuclear envelope tethering and microtubule motor-based pulling forces collaborate in centrosome positioning during mitotic entry Vincent Boudreau1, Richard Chen1, Alan Edwards1, Muhammad Sulaimain1, Paul S. Maddox1* 1 Department of Biology, University of North Carolina at Chapel Hill * Direct all correspondence to this author at [email protected]. Centrosome positioning relative to the nucleus and cell nuclear envelope and at the cortex to ensure proper centrosome shape is highly regulated across cell types, during cell migration positioning (De Simone et al., 2016). Dynein regulators including and during spindle formation in cell division. Across most sexual- LIS-1 are also required for centrosome separation in the embryo ly reproducing animals, centrosomes are provided to the oocyte (Cockell et al., 2004), although their spatio-temporal contributions through fertilization and must be positioned properly to establish remain elusive. Despite our understanding of microtubule cyto- the zygotic mitotic spindle. How centrosomes are positioned in skeleton-based pulling forces, the contribution of key mitotic kinas- space and time through the concerted action of key mitotic en- es and phosphatases to centrosome positioning remains unclear. try biochemical regulators including Protein Phosphatase 2A The net effect of molecular regulators is a biophysical (PP2A-B55/SUR-6), biophysical regulators including Dynein mechanism required for positioning centrosomes during mitotic and the nuclear lamina is unclear. -

Introduction to the Cell Cell History Cell Structures and Functions
Introduction to the cell cell history cell structures and functions CK-12 Foundation December 16, 2009 CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-content, web-based collaborative model termed the “FlexBook,” CK-12 intends to pioneer the generation and distribution of high quality educational content that will serve both as core text as well as provide an adaptive environment for learning. Copyright ©2009 CK-12 Foundation This work is licensed under the Creative Commons Attribution-Share Alike 3.0 United States License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/us/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Contents 1 Cell structure and function dec 16 5 1.1 Lesson 3.1: Introduction to Cells .................................. 5 3 www.ck12.org www.ck12.org 4 Chapter 1 Cell structure and function dec 16 1.1 Lesson 3.1: Introduction to Cells Lesson Objectives • Identify the scientists that first observed cells. • Outline the importance of microscopes in the discovery of cells. • Summarize what the cell theory proposes. • Identify the limitations on cell size. • Identify the four parts common to all cells. • Compare prokaryotic and eukaryotic cells. Introduction Knowing the make up of cells and how cells work is necessary to all of the biological sciences. Learning about the similarities and differences between cell types is particularly important to the fields of cell biology and molecular biology. -

The Nature of Genomes Viral Genomes Prokaryotic Genome
The nature of genomes • Genomics: study of structure and function of genomes • Genome size – variable, by orders of magnitude – number of genes roughly proportional to genome size • Plasmids – symbiotic DNA molecules, not essential – mostly circular in prokaryotes • Organellar DNA – chloroplast, mitochondrion – derived by endosymbiosis from bacterial ancestors Chapter 2: Genes and genomes © 2002 by W. H. Freeman and Company Chapter 2: Genes and genomes © 2002 by W. H. Freeman and Company Viral genomes • Nonliving particle In prokaryotes, viruses are – nucleic acid sometimes referred to as – protein bacteriophages. • DNA or RNA – single-stranded or double-stranded – linear or circular • Compact genomes with little spacer DNA Chapter 2: Genes and genomes © 2002 by W. H. Freeman and Company Chapter 2: Genes and genomes © 2002 by W. H. Freeman and Company Prokaryotic genome • Usually circular double helix – occupies nucleoid region of cell – attached to plasma membrane • Genes are close together with little intergenic spacer • Operon – tandem cluster of coordinately regulated genes – transcribed as single mRNA • Introns very rare Chapter 2: Genes and genomes © 2002 by W. H. Freeman and Company Chapter 2: Genes and genomes © 2002 by W. H. Freeman and Company 1 Eukaryotic nuclear genomes • Each species has characteristic chromosome number • Genes are segments of nuclear chromosomes • Ploidy refers to number of complete sets of chromosomes –haploid (1n): one complete set of genes – diploid (2n) – polyploid (≥3n) • In diploids, chromosomes come in homologous pairs (homologs) In humans, somatic cells have – structurally similar 2n = 46 chromosomes. – same sequence of genes – may contain different alleles Chapter 2: Genes and genomes © 2002 by W. H. -

Principles of Human Anatomy
Principles of Cells Human Anatomy Cells are the basic living structural, Eleventh Edition functional unit of the body Gerard J. Tortora Cytology is the branch of science that & studies cells Mark T. Nielsen The human body has 100 trillion cells 200 CHAPTER 2 different cell types with a variety of Cells shapes, sizes and functions. Copyright © 2007 by John Wiley & Sons, Inc. Cell Diversity Generalized Cell Sizes (diameter) Ovum – 140 µm RBC – 8 µm Major parts of a cell µm = 1/10,000 of a cm Shapes Plasma membrane Flat Cytoplasm Oval Cubed Organelles Star shaped Elongated Inclusions Concave Structures Flagella Cilia Microvilli Fluid mosaic model of the plasma membrane Membrane Lipids Phospholipids – 75% Lipid bilayer Glycolipids – 5% Self recognition Cholesterol – 20% Maintains integrity Maintains fluidity Membrane Proteins Functions of the Cell Membrane Integral proteins Extend across the Communication phospholipid bilayer Shape & protection Channels Pores Maintains the electrochemical gradient Receptors Electrical separation of charge Transporters Enzymes Chemical (concentration gradient) Peripheral proteins Selective permeability Loosely attached to inner or outer surface Some substances easily travel across the Enzymes membrane and others do not Cytoskeletal anchors Membrane Transport Membrane Transport Active transport (uses ATP) Passive transport (kinetic energy not ATP) Primary active transport Net diffusion Molecule mover hydrolyzes ATP Movement of molecules from [high] to [low] -

Centrosome Positioning in Vertebrate Development
Commentary 4951 Centrosome positioning in vertebrate development Nan Tang1,2,*,` and Wallace F. Marshall2,` 1Department of Anatomy, Cardiovascular Research Institute, The University of California, San Francisco, USA 2Department Biochemistry and Biophysics, The University of California, San Francisco, USA *Present address: National Institute of Biological Science, Beijing, China `Authors for correspondence ([email protected]; [email protected]) Journal of Cell Science 125, 4951–4961 ß 2012. Published by The Company of Biologists Ltd doi: 10.1242/jcs.038083 Summary The centrosome, a major organizer of microtubules, has important functions in regulating cell shape, polarity, cilia formation and intracellular transport as well as the position of cellular structures, including the mitotic spindle. By means of these activities, centrosomes have important roles during animal development by regulating polarized cell behaviors, such as cell migration or neurite outgrowth, as well as mitotic spindle orientation. In recent years, the pace of discovery regarding the structure and composition of centrosomes has continuously accelerated. At the same time, functional studies have revealed the importance of centrosomes in controlling both morphogenesis and cell fate decision during tissue and organ development. Here, we review examples of centrosome and centriole positioning with a particular emphasis on vertebrate developmental systems, and discuss the roles of centrosome positioning, the cues that determine positioning and the mechanisms by which centrosomes respond to these cues. The studies reviewed here suggest that centrosome functions extend to the development of tissues and organs in vertebrates. Key words: Centrosome, Development, Mitotic spindle orientation Introduction radiating out to the cell cortex (Fig. 2A). In some cases, the The centrosome of animal cells (Fig. -
Chapter 4 – Cell Structure
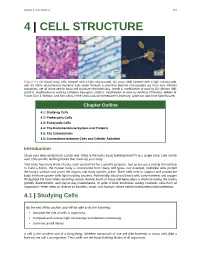
Chapter 4 | Cell Structure 107 4 | CELL STRUCTURE Figure 4.1 (a) Nasal sinus cells (viewed with a light microscope), (b) onion cells (viewed with a light microscope), and (c) Vibrio tasmaniensis bacterial cells (seen through a scanning electron microscope) are from very different organisms, yet all share certain basic cell structure characteristics. (credit a: modification of work by Ed Uthman, MD; credit b: modification of work by Umberto Salvagnin; credit c: modification of work by Anthony D'Onofrio, William H. Fowle, Eric J. Stewart, and Kim Lewis of the Lewis Lab at Northeastern University; scale-bar data from Matt Russell) Chapter Outline 4.1: Studying Cells 4.2: Prokaryotic Cells 4.3: Eukaryotic Cells 4.4: The Endomembrane System and Proteins 4.5: The Cytoskeleton 4.6: Connections between Cells and Cellular Activities Introduction Close your eyes and picture a brick wall. What is the wall's basic building block? It is a single brick. Like a brick wall, cells are the building blocks that make up your body. Your body has many kinds of cells, each specialized for a specific purpose. Just as we use a variety of materials to build a home, the human body is constructed from many cell types. For example, epithelial cells protect the body's surface and cover the organs and body cavities within. Bone cells help to support and protect the body. Immune system cells fight invading bacteria. Additionally, blood and blood cells carry nutrients and oxygen throughout the body while removing carbon dioxide. Each of these cell types plays a vital role during the body's growth, development, and day-to-day maintenance. -

Introduction
Oncogene (2002) 21, 6140 – 6145 ª 2002 Nature Publishing Group All rights reserved 0950 – 9232/02 $25.00 www.nature.com/onc Introduction Kenji Fukasawa*,1 1Department of Cell Biology, University of Cincinnati College of Medicine, PO Box 670521, Cincinnati, Ohio, OH 45267-0521, USA Oncogene (2002) 21, 6140 – 6145. doi:10.1038/sj.onc. Centrosomes have recently attracted considerable 1205771 attention primarily because of their potential impor- tance in carcinogenesis. Chromosome instability is a hallmark of virtually all solid cancers, being a Keywords: centrosome; cancer; chromosome instability formidable force that drives multi-step carcinogenesis: either loss or gain of a single chromosome can simultaneously introduce multiple mutations, which are responsible for acquisition of further malignant The centrosome of animal cells is a small non- phenotypes. The presence of more than two centro- membranous organelle, and is often associated with somes in a cell results in the formation of defective the nuclear membrane. It is composed of a pair of mitotic spindles directed by multiple spindle poles, centrioles and a surrounding electron dense matrix of which in turn increases the chromosome segregation protein aggregates referred to as the pericentriolar errors. This potential role of centrosomes in chromo- material (PCM) (Figure 1, also see Figure 1 in Dutertre some instability, hence in cancer development, is by et al., 2002). Each centriole is cylindrical in shape and no means a new-sprung idea. It was initially built with the nine sets of triplet microtubules. The two proposed by Theodor Boveri (1914). In his book, centrioles are paired in close proximity at one end, and The Origin of Malignant Tumors, he wrote, ‘malig- positioned vertical to each other. -

The Emerging Role of Ncrnas and RNA-Binding Proteins in Mitotic Apparatus Formation
non-coding RNA Review The Emerging Role of ncRNAs and RNA-Binding Proteins in Mitotic Apparatus Formation Kei K. Ito, Koki Watanabe and Daiju Kitagawa * Department of Physiological Chemistry, Graduate School of Pharmaceutical Science, The University of Tokyo, Bunkyo, Tokyo 113-0033, Japan; [email protected] (K.K.I.); [email protected] (K.W.) * Correspondence: [email protected] Received: 11 November 2019; Accepted: 13 March 2020; Published: 20 March 2020 Abstract: Mounting experimental evidence shows that non-coding RNAs (ncRNAs) serve a wide variety of biological functions. Recent studies suggest that a part of ncRNAs are critically important for supporting the structure of subcellular architectures. Here, we summarize the current literature demonstrating the role of ncRNAs and RNA-binding proteins in regulating the assembly of mitotic apparatus, especially focusing on centrosomes, kinetochores, and mitotic spindles. Keywords: ncRNA; centrosome; kinetochore; mitotic spindle 1. Introduction Non-coding RNAs (ncRNAs) are defined as a class of RNA molecules that are transcribed from genomic DNA, but not translated into proteins. They are mainly classified into the following two categories according to their length—small RNA (<200 nt) and long non-coding RNA (lncRNA) (>200 nt). Small RNAs include traditional RNA molecules, such as transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), PIWI-interacting RNA (piRNA), and micro RNA (miRNA), and they have been studied extensively [1]. Research on lncRNA is behind that on small RNA despite that recent transcriptome analysis has revealed that more than 120,000 lncRNAs are generated from the human genome [2–4]. -

The Mitochondrial Genome. the Nucleoid
ISSN 0006-2979, Biochemistry (Moscow), 2016, Vol. 81, No. 10, pp. 1057-1065. © Pleiades Publishing, Ltd., 2016. Original Russian Text © A. A. Kolesnikov, 2016, published in Biokhimiya, 2016, Vol. 81, No. 10, pp. 1322-1331. REVIEW The Mitochondrial Genome. The Nucleoid A. A. Kolesnikov Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia; E-mail: [email protected] Received May 30, 2016 Revision received July 1, 2016 Abstract—Mitochondrial DNA (mtDNA) in cells is organized in nucleoids containing DNA and various proteins. This review discusses questions of organization and structural dynamics of nucleoids as well as their protein components. The structures of mt-nucleoid from different organisms are compared. The currently accepted model of nucleoid organization is described and questions needing answers for better understanding of the fine mechanisms of the mitochondrial genetic apparatus functioning are discussed. DOI: 10.1134/S0006297916100047 Key words: mitochondrial nucleoid, mitochondrial genome It is now clear that understanding of genome organ- mtDNA). In fact, proteins associated with a nucleoid can ization of subcellular structures is necessary for under- influence the speed of accumulation of mutations. standing of cellular processes occurring through the cell In the last decade, reviews devoted to this subject cycle, including questions of storage, realization, and appeared regularly [5-9]. Generally, they cover the subject transmission of genetic information. A considerable of nucleoid organization in metazoan mitochondria amount of information about forms and sizes of mito- (mainly in human cells) and fungi (mainly yeast). chondrial DNA (mtDNA) throughout evolution has been Reviews touching plant cells are much less frequent [10, accumulated [1]. -

The Central Role of Centrosomes in Translocation of Mitochondria and Centrosome Proteins
916 Microsc Microanal 11(Suppl 2), 2005 DOI: 10.1017/S1431927605505798 Copyright 2005 Microscopy Society of America The central role of centrosomes in translocation of mitochondria and centrosome proteins H. Schatten*, Z.H. Liu**, Y.H Hao**, L.X Lai**, D.Wax**, M. Samuel**, Q.-Y. Sun***, and R.S. Prather** *Dept. of Veterinary Pathobiology, University of Missouri-Columbia, Columbia, MO 65211 **Division of Animal Science, University of Missouri-Columbia, Columbia MO 65211 ***State Key Laboratory of Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100080, P.R. China Centrosomes are central cell bodies that are predominantly known for their role as microtubule organizing centers. However, by nucleating and organizing microtubules into specific patterns, centrosomes play also a crucial role in directing the translocation of cell organelles, vesicles, and proteins to their functional destinations. Here we review translocation of mitochondria and the nuclear mitotic apparatus protein (NuMA) in somatic cells and during oocyte maturation, parthenogenetic activation, fertilization, nuclear cloning, and development in porcine oocytes. Our studies have shown that a temporal, spatial, and developmental relationship exists between microtubule organization and the progressive translocation of mitochondria and NuMA allowing dynamic and precisely coordinated cell and molecular interactions. During normal fertilization in most species centrosomes are crucial for the organization of the sperm aster that unites maternal and paternal genomes during fertilization and for the equal distribution of DNA and cell organelles during cell division, cell differentiation, and development [1,2,3,4,5,6]. Mitochondria play a significant role in cellular metabolism by providing energy in form of ATP that is required for specific cell cycle and developmental events. -
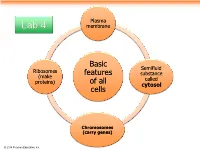
Basic Features of All Cells
Plasma Lab 4 membrane Basic Semifluid Ribosomes features substance (make called proteins) of all cytosol cells Chromosomes (carry genes) © 2014 Pearson Education, Inc. Prokaryotic cells are characterized by having . No nucleus . DNA in an unbound region called the nucleoid . No membrane-bound organelles . Cytoplasm bound by the plasma membrane © 2014 Pearson Education, Inc. Eukaryotic cells are characterized by having • DNA in a nucleus that is bounded by a membranous nuclear envelope • Membrane-bound organelles • Cytoplasm in the region between the plasma membrane and nucleus Eukaryotic cells are generally much larger than prokaryotic cells © 2014 Pearson Education, Inc. Figure 6.8a ENDOPLASMIC RETICULUM (ER) Nuclear envelope Rough ER Smooth ER Nucleolus NUCLEUS Flagellum Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes Microvilli Golgi apparatus Peroxisome Lysosome Mitochondrion © 2014 Pearson Education, Inc. Figure 6.8b Nuclear envelope NUCLEUS Nucleolus Rough ER Chromatin Smooth ER Ribosomes Golgi Central vacuole apparatus Microfilaments CYTOSKELETON Microtubules Mitochondrion Peroxisome Plasma Chloroplast membrane Cell wall Plasmodesmata Wall of adjacent cell © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. Table 6.1 © 2014 Pearson Education, Inc. Cell Walls of Plants The cell wall is an extracellular structure that distinguishes plant cells from animal cells Plant cell walls Prokaryotes, are made