NGS Barcoding Reveals High Resistance of a Hyperdiverse Chironomid (Diptera) Swamp Fauna Against Invasion from Adjacent Freshwat
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Taxonomic Review of the Chironomid Genus Cricotopus V.D. Wulp
Zootaxa 3919 (1): 001–040 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2015 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3919.1.1 http://zoobank.org/urn:lsid:zoobank.org:pub:218630EE-6BF7-4E35-A8F6-9E8260D60FA0 Taxonomic review of the chironomid genus Cricotopus v.d. Wulp (Diptera: Chironomidae) from Australia: keys to males, females, pupae and larvae, description of ten new species and comments on Paratrichocladius Santos Abreu NICK DRAYSON1, PETER S. CRANSTON2,4 & MATT N. KROSCH3 17 Park Walk, Brigstock, Northants NN14 3HH, UK 2Evolution, Ecology and Genetics, Research School of Biology, The Australian National University, Canberra, A.C.T. 2601, Australia 3Centre for Water in the Minerals Industry, Sustainable Minerals Institute, The University of Queensland., Brisbane, QLD 4072, Australia 4Corresponding author Drayson: urn:lsid:zoobank.org:author:B568A060-A52A-4440-8CC1-D81506B3902A Cranston: urn:lsid:zoobank.org:author:C068AC61-DF1D-432A-9AB7-52B5D85C6C79 Krosch: urn:lsid:zoobank.org:author:C7DD7291-27F0-4216-80B2-90BD9F0CDFAB Table of contents Abstract . 1 Introduction . 2 Methods and material . 3 Cricotopus acornis Drayson & Cranston, sp.n. 5 Cricotopus albitarsis Hergstrom sp. n. 6 Cricotopus annuliventris (Skuse) . 8 Cricotopus brevicornis Drayson & Cranston sp.n. 10 Cricotopus conicornis Drayson & Cranston sp.n.. 11 Cricotopus hillmani Drayson & Cranston, sp. n . 13 Cricotopus howensis Cranston sp.n. 15 Cricotopus parbicinctus Hergstrom sp.n. 16 Cricotopus tasmania Drayson & Cranston sp. n. 18 Cricotopus varicornis Drayson & Cranston sp. n. 20 Cricotopus wangi Cranston & Krosch sp. n.. 21 Key to adult males of Australian Cricotopus . 22 Key to adult females of Australian Cricotopus . -

Distribution of Freshwater Midges in Relation to Air Temperature and Lake Depth
J Paleolimnol (2006) 36:295-314 DOl 1O.1007/s10933-006-0014-6 A northwest North American training set: distribution of freshwater midges in relation to air temperature and lake depth Erin M. Barley' Ian R. Walker' Joshua Kurek, Les C. Cwynar' Rolf W. Mathewes • Konrad Gajewski' Bruce P. Finney Received: 20 July 2005 I Accepted: 5 March 2006/Published online: 26 August 2006 © Springer Science+Business Media B.Y. 2006 Abstract Freshwater midges, consisting of Chiro- organic carbon, lichen woodland vegetation and sur- nomidae, Chaoboridae and Ceratopogonidae, were face area contributed significantly to explaining assessed as a biological proxy for palaeoclimate in midge distribution. Weighted averaging partial least eastern Beringia. The northwest North American squares (WA-PLS) was used to develop midge training set consists of midge assemblages and data inference models for mean July air temperature 2 for 17 environmental variables collected from 145 (R boot = 0.818, RMSEP = 1.46°C), and transformed 2 lakes in Alaska, British Columbia, Yukon, Northwest depth (1n (x+ l); R boot = 0.38, and RMSEP = 0.58). Territories, and the Canadian Arctic Islands. Canon- ical correspondence analyses (CCA) revealed that Key words Chironomidae : Transfer function . mean July air temperature, lake depth, arctic tundra Beringia' Air temperature . Lake depth' Canonical vegetation, alpine tundra vegetation, pH, dissolved correspondence analysis . Paleoclimate E. M. Barley· R. W. Mathewes Department of Biological Sciences, Simon Fraser University, Burnaby, British Columbia, Canada V5A IS6 Introduction I. R. Walker Departments of Biology, and Earth and Environmental Sciences, University of British Columbia Okanagan, Palaeoecologists seeking to quantify past environ- Kelowna, BC, Canada VIV IV7 mental changes rely increasingly on transfer func- e-mail: [email protected] tions that make use of biological proxies (Battarbee J. -

Orthocladiinae 7.1
ORTHOCLADIINAE 7.1 SUBFAMILY ORTHOCLADIINAE 7 DIAGNOSIS: Antennae with 3-7 segments; may be strongly reduced or may be longer than head capsule. Labrum with S I variable (simple, bifid, branched, serrated, palmate or plumose); S II usually simple but may be bifid, branched, palmate or plumose; S III simple (rarely bifid); S IV normal. Labral lamellae present or absent. Mentum usually well sclerotized, with several to more than 25 teeth; ventro- mental plates absent/vestigial to very large, without striae (occasionally with ridges in Nanocladius); beard present or absent. Prementum variably developed but never with dense well developed median brush of setae. Body with anterior parapods (sometimes reduced and/or fused); with posterior parapods well developed, separate or fused, or parapods reduced or absent. Setal fringe, setal tufts or long setae sometimes present. Anal tubules normally present, may be reduced or absent/vestigial. NOTES: One of the most diverse of the chironomid subfamilies; orthoclad larvae are found in an amaz- ing variety of habitats, running the gamut from terrestrial (corn fields, dung, greenhouses, leaf litter in hardwood forests) to seeps, springs, streams, rivers, ponds and lakes in freshwater, and coastal estuarine and littoral marine areas. Most larvae are scrapers, shredders or collectors-gatherers; some taxa are preda- tors, some are parasites. Key to the genera of larval Orthocladiinae of the southeastern United States (larvae are unknown for Apometriocnemus, Chasmatonotus, Diplosmittia, Lipurometriocnemus, Plhudsonia, Saetheriella, Sublettiella and Tavastia) 1 Length of antennae at least 1/2 length of head capsule ............................................................ 2 1’ Length of antennae less than 1/2 length of head capsule ........................................................ -

Chironominae 8.1
CHIRONOMINAE 8.1 SUBFAMILY CHIRONOMINAE 8 DIAGNOSIS: Antennae 4-8 segmented, rarely reduced. Labrum with S I simple, palmate or plumose; S II simple, apically fringed or plumose; S III simple; S IV normal or sometimes on pedicel. Labral lamellae usually well developed, but reduced or absent in some taxa. Mentum usually with 8-16 well sclerotized teeth; sometimes central teeth or entire mentum pale or poorly sclerotized; rarely teeth fewer than 8 or modified as seta-like projections. Ventromental plates well developed and usually striate, but striae reduced or vestigial in some taxa; beard absent. Prementum without dense brushes of setae. Body usually with anterior and posterior parapods and procerci well developed; setal fringe not present, but sometimes with bifurcate pectinate setae. Penultimate segment sometimes with 1-2 pairs of ventral tubules; antepenultimate segment sometimes with lateral tubules. Anal tubules usually present, reduced in brackish water and marine taxa. NOTESTES: Usually the most abundant subfamily (in terms of individuals and taxa) found on the Coastal Plain of the Southeast. Found in fresh, brackish and salt water (at least one truly marine genus). Most larvae build silken tubes in or on substrate; some mine in plants, dead wood or sediments; some are free- living; some build transportable cases. Many larvae feed by spinning silk catch-nets, allowing them to fill with detritus, etc., and then ingesting the net; some taxa are grazers; some are predacious. Larvae of several taxa (especially Chironomus) have haemoglobin that gives them a red color and the ability to live in low oxygen conditions. With only one exception (Skutzia), at the generic level the larvae of all described (as adults) southeastern Chironominae are known. -

Checklist of the Family Chironomidae (Diptera) of Finland
A peer-reviewed open-access journal ZooKeys 441: 63–90 (2014)Checklist of the family Chironomidae (Diptera) of Finland 63 doi: 10.3897/zookeys.441.7461 CHECKLIST www.zookeys.org Launched to accelerate biodiversity research Checklist of the family Chironomidae (Diptera) of Finland Lauri Paasivirta1 1 Ruuhikoskenkatu 17 B 5, FI-24240 Salo, Finland Corresponding author: Lauri Paasivirta ([email protected]) Academic editor: J. Kahanpää | Received 10 March 2014 | Accepted 26 August 2014 | Published 19 September 2014 http://zoobank.org/F3343ED1-AE2C-43B4-9BA1-029B5EC32763 Citation: Paasivirta L (2014) Checklist of the family Chironomidae (Diptera) of Finland. In: Kahanpää J, Salmela J (Eds) Checklist of the Diptera of Finland. ZooKeys 441: 63–90. doi: 10.3897/zookeys.441.7461 Abstract A checklist of the family Chironomidae (Diptera) recorded from Finland is presented. Keywords Finland, Chironomidae, species list, biodiversity, faunistics Introduction There are supposedly at least 15 000 species of chironomid midges in the world (Armitage et al. 1995, but see Pape et al. 2011) making it the largest family among the aquatic insects. The European chironomid fauna consists of 1262 species (Sæther and Spies 2013). In Finland, 780 species can be found, of which 37 are still undescribed (Paasivirta 2012). The species checklist written by B. Lindeberg on 23.10.1979 (Hackman 1980) included 409 chironomid species. Twenty of those species have been removed from the checklist due to various reasons. The total number of species increased in the 1980s to 570, mainly due to the identification work by me and J. Tuiskunen (Bergman and Jansson 1983, Tuiskunen and Lindeberg 1986). -

Flies) Benjamin Kongyeli Badii
Chapter Phylogeny and Functional Morphology of Diptera (Flies) Benjamin Kongyeli Badii Abstract The order Diptera includes all true flies. Members of this order are the most ecologically diverse and probably have a greater economic impact on humans than any other group of insects. The application of explicit methods of phylogenetic and morphological analysis has revealed weaknesses in the traditional classification of dipteran insects, but little progress has been made to achieve a robust, stable clas- sification that reflects evolutionary relationships and morphological adaptations for a more precise understanding of their developmental biology and behavioral ecol- ogy. The current status of Diptera phylogenetics is reviewed in this chapter. Also, key aspects of the morphology of the different life stages of the flies, particularly characters useful for taxonomic purposes and for an understanding of the group’s biology have been described with an emphasis on newer contributions and progress in understanding this important group of insects. Keywords: Tephritoidea, Diptera flies, Nematocera, Brachycera metamorphosis, larva 1. Introduction Phylogeny refers to the evolutionary history of a taxonomic group of organisms. Phylogeny is essential in understanding the biodiversity, genetics, evolution, and ecology among groups of organisms [1, 2]. Functional morphology involves the study of the relationships between the structure of an organism and the function of the various parts of an organism. The old adage “form follows function” is a guiding principle of functional morphology. It helps in understanding the ways in which body structures can be used to produce a wide variety of different behaviors, including moving, feeding, fighting, and reproducing. It thus, integrates concepts from physiology, evolution, anatomy and development, and synthesizes the diverse ways that biological and physical factors interact in the lives of organisms [3]. -

Flies of the Nearctic Region
Eur. J. Entomol. 102: 298, 2005 ISSN 1210-5759 BOOK REVIEW HALL J.C. & EVENHUIS N.L.: Homeodactyla and Asilomorpha. In The arrangement of the systematic part follows the usual and Griffiths G.C.D. (ed.): FLIES OF THE NEARCTIC REGION. well-established organisation of the whole series “Flies of the Vol. V, Part 13, No. 7, Bombyliidae. pp. 657–713. E. Schweiz- Nearctic Region”. Each genus is headed by a full synonymy erbart’sche Verlagsbuchhandlung, Stuttgart, 2004, 60 pp., 46 with a history of its classification and an accepted or proposed Figs. ISBN 3-510-70027-9. Price EUR 39.00. classification into groups of species, followed by chapters on zoogeography, morphological diagnosis and notes on biology. This is the concluding part of an elaborate account of the Each species is fully described, the main differential features are family Bombyliidae (Diptera) in the series Flies of the Nearctic illustrated (mainly antennae and male genitalia), and brief notes Region, edited by the leading Canadian dipterist Graham C.D. on variation, differential diagnoses and life histories are also Griffiths. This already worldwide famous North American added. Distribution of each species is based on full data from series is published in Europe, in Stuttgart, Germany. The first the revised specimens or types. A key to Nearctic species is six parts on the Bombyliidae appeared between 1980 and 1987, always given at the end of each genus, and cited literature at the but the 7th, and concluding part, is only now available after a end of each subfamily. This concluding part of the series on delay of 17 years. -

Bibliograp Bibliography
BIBLIOGRAPHY 9.1 BIBLIOGRAPHY 9 Adam, J.I & O.A. Sæther. 1999. Revision of the records for the southern United States genus Nilothauma Kieffer, 1921 (Diptera: (Diptera: Chironomidae). J. Ga. Ent. Soc. Chironomidae). Ent. scand. Suppl. 56: 1- 15:69-73. 107. Beck, W.M., Jr. & E.C. Beck. 1966. Chironomidae Ali, A. 1991. Perspectives on management of pest- (Diptera) of Florida - I. Pentaneurini (Tany- iferous Chironomidae (Diptera), an emerg- podinae). Bull. Fla. St. Mus. Biol. Sci. ing global problem. J. Am. Mosq. Control 10:305-379. Assoc. 7: 260-281. Beck, W.M., Jr., & E.C. Beck. 1970. The immature Armitage, P., P.S. Cranston & L.C.V. Pinder (eds). stages of some Chironomini (Chiro- 1995. The Chironomidae. Biology and nomidae). Q.J. Fla. Acad. Sci. 33:29-42. ecology of non-biting midges. Chapman & Bilyj, B. 1984. Descriptions of two new species of Hall, London. 572 pp. Tanypodinae (Diptera: Chironomidae) from Ashe, P. 1983. A catalogue of chironomid genera Southern Indian Lake, Canada. Can. J. Fish. and subgenera of the world including syn- Aquat. Sci. 41: 659-671. onyms (Diptera: Chironomidae). Ent. Bilyj, B. 1985. New placement of Tanypus pallens scand. Suppl. 17: 1-68. Coquillett, 1902 nec Larsia pallens (Coq.) Barton, D.R., D.R. Oliver & M.E. Dillon. 1993. sensu Roback 1971 (Diptera: Chironomi- The first Nearctic record of Stackelbergina dae) and redescription of the holotype. Can. Shilova and Zelentsov (Diptera: Chironomi- Ent. 117: 39-42. dae): Taxonomic and ecological observations. Bilyj, B. 1988. A taxonomic review of Guttipelopia Aquatic Insects 15: 57-63. (Diptera: Chironomidae). -

DNA Barcoding
Full-time PhD studies of Ecology and Environmental Protection Piotr Gadawski Species diversity and origin of non-biting midges (Chironomidae) from a geologically young lake PhD Thesis and its old spring system Performed in Department of Invertebrate Zoology and Hydrobiology in Institute of Ecology and Environmental Protection Różnorodność gatunkowa i pochodzenie fauny Supervisor: ochotkowatych (Chironomidae) z geologicznie Prof. dr hab. Michał Grabowski młodego jeziora i starego systemu źródlisk Auxiliary supervisor: Dr. Matteo Montagna, Assoc. Prof. Łódź, 2020 Łódź, 2020 Table of contents Acknowledgements ..........................................................................................................3 Summary ...........................................................................................................................4 General introduction .........................................................................................................6 Skadar Lake ...................................................................................................................7 Chironomidae ..............................................................................................................10 Species concept and integrative taxonomy .................................................................12 DNA barcoding ...........................................................................................................14 Chapter I. First insight into the diversity and ecology of non-biting midges (Diptera: Chironomidae) -
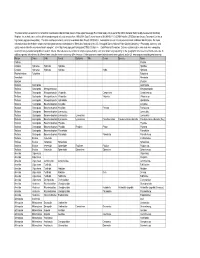
This Table Contains a Taxonomic List of Benthic Invertebrates Collected from Streams in the Upper Mississippi River Basin Study
This table contains a taxonomic list of benthic invertebrates collected from streams in the Upper Mississippi River Basin study unit as part of the USGS National Water Quality Assessemnt (NAWQA) Program. Invertebrates were collected from woody snags in selected streams from 1996-2004. Data Retreival occurred 26-JAN-06 11.10.25 AM from the USGS data warehouse (Taxonomic List Invert http://water.usgs.gov/nawqa/data). The data warehouse currently contains invertebrate data through 09/30/2002. Invertebrate taxa can include provisional and conditional identifications. For more information about invertebrate sample processing and taxonomic standards see, "Methods of analysis by the U.S. Geological Survey National Water Quality Laboratory -- Processing, taxonomy, and quality control of benthic macroinvertebrate samples", at << http://nwql.usgs.gov/Public/pubs/OFR00-212.html >>. Data Retrieval Precaution: Extreme caution must be exercised when comparing taxonomic lists generated using different search criteria. This is because the number of samples represented by each taxa list will vary depending on the geographic criteria selected for the retrievals. In addition, species lists retrieved at different times using the same criteria may differ because: (1) the taxonomic nomenclature (names) were updated, and/or (2) new samples containing new taxa may Phylum Class Order Family Subfamily Tribe Genus Species Taxon Porifera Porifera Cnidaria Hydrozoa Hydroida Hydridae Hydridae Cnidaria Hydrozoa Hydroida Hydridae Hydra Hydra sp. Platyhelminthes Turbellaria Turbellaria Nematoda Nematoda Bryozoa Bryozoa Mollusca Gastropoda Gastropoda Mollusca Gastropoda Mesogastropoda Mesogastropoda Mollusca Gastropoda Mesogastropoda Viviparidae Campeloma Campeloma sp. Mollusca Gastropoda Mesogastropoda Viviparidae Viviparus Viviparus sp. Mollusca Gastropoda Mesogastropoda Hydrobiidae Hydrobiidae Mollusca Gastropoda Basommatophora Ancylidae Ancylidae Mollusca Gastropoda Basommatophora Ancylidae Ferrissia Ferrissia sp. -

Surveying for Terrestrial Arthropods (Insects and Relatives) Occurring Within the Kahului Airport Environs, Maui, Hawai‘I: Synthesis Report
Surveying for Terrestrial Arthropods (Insects and Relatives) Occurring within the Kahului Airport Environs, Maui, Hawai‘i: Synthesis Report Prepared by Francis G. Howarth, David J. Preston, and Richard Pyle Honolulu, Hawaii January 2012 Surveying for Terrestrial Arthropods (Insects and Relatives) Occurring within the Kahului Airport Environs, Maui, Hawai‘i: Synthesis Report Francis G. Howarth, David J. Preston, and Richard Pyle Hawaii Biological Survey Bishop Museum Honolulu, Hawai‘i 96817 USA Prepared for EKNA Services Inc. 615 Pi‘ikoi Street, Suite 300 Honolulu, Hawai‘i 96814 and State of Hawaii, Department of Transportation, Airports Division Bishop Museum Technical Report 58 Honolulu, Hawaii January 2012 Bishop Museum Press 1525 Bernice Street Honolulu, Hawai‘i Copyright 2012 Bishop Museum All Rights Reserved Printed in the United States of America ISSN 1085-455X Contribution No. 2012 001 to the Hawaii Biological Survey COVER Adult male Hawaiian long-horned wood-borer, Plagithmysus kahului, on its host plant Chenopodium oahuense. This species is endemic to lowland Maui and was discovered during the arthropod surveys. Photograph by Forest and Kim Starr, Makawao, Maui. Used with permission. Hawaii Biological Report on Monitoring Arthropods within Kahului Airport Environs, Synthesis TABLE OF CONTENTS Table of Contents …………….......................................................……………...........……………..…..….i. Executive Summary …….....................................................…………………...........……………..…..….1 Introduction ..................................................................………………………...........……………..…..….4 -

Diptera) in Delineating Ecological Classifications of Missouri Streams at Different Environmental Scales
Congruence between nutrient water quality parameters and Chironomidae (Diptera) in delineating ecological classifications of Missouri streams at different environmental scales. Kansas Biological Survey Report No. 159 Dec. 2009 by Barbara Hayford*, Donald Huggins†, Debra Baker†, and Michael Johnson‡ †Central Plains Center for BioAssessment Kansas Biological Survey University of Kansas, Lawrence, KS, U.S.A. *Department of Life Sciences Wayne State College, Wayne, NE, U.S.A. ‡Aquatic Ecosystems Analysis Laboratory John Muir Institute of the Environment University of California, Davis, CA, U.S.A. 1 SUMMARY 1. Large scale classifications such as nutrient ecoregions have been established in the United States to help regulate surface water resources. These classifications and their constituent classes are hypotheses which may be tested by establishing the strength of the classes to differentiate within class and between class variation. The purpose of our study was to test the strength of three different classification schemes at three different scales: landscape (nutrient ecoregion), watershed (hydrologic unit), and microhabitat (substrate) using Chironomidae community assemblage and metric data and nutrient water quality data. Furthermore, we tested for congruence of classes using nutrient water quality and chironomid community data for the three classification schemes in an attempt to link the biota to nutrient concentrations. 2. Historical data on chironomid communities were compiled from the 20 stream sites in Missouri over multiple years and seasons. Nutrient water quality data were also collected at these sites but under a different study. An analysis of variance was used to determine whether chironomid metrics and nutrient water quality parameters varied significantly between classes for each of the classification schemes.