The Histone Demethylase KDM3A Regulates the Transcriptional Program of the Androgen Receptor in Prostate Cancer Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

KLF2 Induced
UvA-DARE (Digital Academic Repository) The transcription factor KLF2 in vascular biology Boon, R.A. Publication date 2008 Link to publication Citation for published version (APA): Boon, R. A. (2008). The transcription factor KLF2 in vascular biology. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:23 Sep 2021 Supplementary data: Genes induced by KLF2 Dekker et al. LocusLink Accession Gene Sequence Description Fold p-value ID number symbol change (FDR) 6654 AK022099 SOS1 cDNA FLJ12037 fis, clone HEMBB1001921. 100.00 5.9E-09 56999 AF086069 ADAMTS9 full length insert cDNA clone YZ35C05. 100.00 1.2E-09 6672 AF085934 SP100 full length insert cDNA clone YR57D07. 100.00 6.7E-13 9031 AF132602 BAZ1B Williams Syndrome critical region WS25 mRNA, partial sequence. -

Angiogenic Patterning by STEEL, an Endothelial-Enriched Long
Angiogenic patterning by STEEL, an endothelial- enriched long noncoding RNA H. S. Jeffrey Mana,b, Aravin N. Sukumara,b, Gabrielle C. Lamc,d, Paul J. Turgeonb,e, Matthew S. Yanb,f, Kyung Ha Kub,e, Michelle K. Dubinskya,b, J. J. David Hob,f, Jenny Jing Wangb,e, Sunit Dasg,h, Nora Mitchelli, Peter Oettgeni, Michael V. Seftonc,d,j, and Philip A. Marsdena,b,e,f,1 aInstitute of Medical Science, University of Toronto, Toronto, ON M5S 1A8, Canada; bKeenan Research Centre for Biomedical Science in the Li Ka Shing Knowledge Institute, St. Michael’s Hospital, University of Toronto, Toronto, ON M5B 1T8, Canada; cDonnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, ON M5S 3E2, Canada; dInstitute of Biomaterials and Biomedical Engineering, University of Toronto, Toronto, ON M5S 3G9, Canada; eDepartment of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON M5S 1A8, Canada; fDepartment of Medical Biophysics, University of Toronto, Toronto, ON M5G 1L7, Canada; gArthur and Sonia Labatt Brain Tumour Research Institute, Hospital for SickKids, University of Toronto, Toronto, ON M5G 1X8, Canada; hDivision of Neurosurgery and Keenan Research Centre for Biomedical Science, St. Michael’s Hospital, University of Toronto, Toronto, ON M5B 1W8, Canada; iDepartment of Medicine, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02115; and jDepartment of Chemical Engineering and Applied Chemistry, University of Toronto, Toronto, ON M5S 3E5, Canada Edited by Napoleone Ferrara, University of California, San Diego, La Jolla, CA, and approved January 24, 2018 (received for review August 28, 2017) Endothelial cell (EC)-enriched protein coding genes, such as endothelial formation in vitro and blood vessel formation in vivo. -

Inhibition of Adipocyte Differentiation by Rorα
FEBS Letters 583 (2009) 2031–2036 journal homepage: www.FEBSLetters.org Inhibition of adipocyte differentiation by RORa Hélène Duez a,b,c,1, Christian Duhem a,b,c,1, Saara Laitinen a,b,c, Prashant S. Patole a,b,c, Mouaadh Abdelkarim a,b,c, Brigitte Bois-Joyeux d, Jean-Louis Danan d, Bart Staels a,b,c,* a Institut Pasteur de Lille, Département d’Athérosclérose, Lille F-59019, France b INSERM UMR 545, Lille F-59019, France c Université Lille Nord de France, Faculté des Sciences Pharmaceutiques et Biologiques et Faculté de Médecine, Lille F-59006, France d Centre National de la Recherche Scientifique FRE3210, Faculté de Médecine René Descartes Paris 5, 75015 Paris, France article info abstract Article history: Here we show that gene expression of the nuclear receptor RORa is induced during adipogenesis, Received 8 April 2009 with RORa4 being the most abundantly expressed isoform in human and murine adipose tissue. Revised 27 April 2009 Over-expression of RORa4 in 3T3-L1 cells impairs adipogenesis as shown by the decreased expres- Accepted 8 May 2009 sion of adipogenic markers and lipid accumulation, accompanied by decreased free fatty acid and Available online 18 May 2009 glucose uptake. By contrast, mouse embryonic fibroblasts from staggerer mice, which carry a muta- Edited by Robert Barouki tion in the RORa gene, differentiate more efficiently into mature adipocytes compared to wild-type cells, a phenotype which is reversed by ectopic RORa4 restoration. Ó 2009 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved. Keywords: Adipogenesis RORa Nuclear receptors RAR-related orphan receptors (RORs) 1. -

Systematic Knockdown of Epigenetic Enzymes Identifies a Novel Histone Demethylase PHF8 Overexpressed in Prostate Cancer with An
Oncogene (2012) 31, 3444–3456 & 2012 Macmillan Publishers Limited All rights reserved 0950-9232/12 www.nature.com/onc ORIGINAL ARTICLE Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion M Bjo¨rkman1,PO¨stling1,2,VHa¨rma¨1, J Virtanen1, J-P Mpindi1,2, J Rantala1,4, T Mirtti2, T Vesterinen2, M Lundin2, A Sankila2, A Rannikko3, E Kaivanto1, P Kohonen1, O Kallioniemi1,2,5 and M Nees1,5 1Medical Biotechnology, VTT Technical Research Centre of Finland, and Center for Biotechnology, University of Turku and A˚bo Akademi University, Turku, Finland; 2Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland and 3HUSLAB, Department of Urology, University of Helsinki, Helsinki, Finland Our understanding of key epigenetic regulators involved in finger proteins like PHF8, are activated in subsets of specific biological processes and cancers is still incom- PrCa’s and promote cancer relevant phenotypes. plete, despite great progress in genome-wide studies of the Oncogene (2012) 31, 3444–3456; doi:10.1038/onc.2011.512; epigenome. Here, we carried out a systematic, genome- published online 28 November 2011 wide analysis of the functional significance of 615 epigenetic proteins in prostate cancer (PrCa) cells. We Keywords: prostate cancer; epigenetics; histone used the high-content cell-spot microarray technology and demethylase; migration; invasion; PHF8 siRNA silencing of PrCa cell lines for functional screening of cell proliferation, survival, androgen receptor (AR) expression, histone methylation and acetylation. Our study highlights subsets of epigenetic enzymes influencing different cancer cell phenotypes. -

Regulation of T Follicular Helper Cells by ICOS
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No. 26 Editorial Regulation of T follicular helper cells by ICOS Andreas Hutloff T follicular helper (TFH) cells are gatekeepers of the (OPN-i), which facilitates nuclear translocation [3]. In the humoral immune response. Without help from this CD4+ nucleus, OPN-i dimerizes with Bcl-6 and protects it from T cell subset, B cells cannot differentiate into high-affinity proteasomal degradation. However, this OPN-i and the memory B cells and antibody-producing long-lived plasma above Klf2 pathway do act at different times. Upon ICOS cells which are the basis of protective immune responses. signaling blockade, Klf2 is upregulated within a few hours At the same time, dysregulated TFH cell responses are and TFH cells lose their typical homing receptor pattern in causative for many autoimmune disorders (reviewed in less than 24 hours [6]. In contrast, complete degradation [1]). The inducible T cell costimulator ICOS, which is of Bcl-6 takes several days [3, 6]. Therefore, the ICOS - structurally and functionally related to CD28, has been Klf2 axis first leads to emigration of TFH cells out of the known for many years as an important regulator of TFH B cell follicle (and thereby to a loss of function), whereas cells. ICOS knock-out mice as well as ICOS-deficient the osteopontin pathway acts as a second strike pathway patients have only few TFH cells and very small germinal eliminating the lineage-defining transcription factor Bcl-6. centers upon immunization, which results in severely Another important finding especially for potential compromised antigen-specific immunoglobulin levels therapeutic applications is that ICOS and the structurally and the phenotype of common variable immunodeficiency related CD28 molecule act in different phases of TFH cell in humans (reviewed in [2]). -

Beta Cell Adaptation to Pregnancy Requires Prolactin Action on Both
www.nature.com/scientificreports OPEN Beta cell adaptation to pregnancy requires prolactin action on both beta and non‑beta cells Vipul Shrivastava1, Megan Lee1, Daniel Lee1, Marle Pretorius1, Bethany Radford1, Guneet Makkar1 & Carol Huang1,2,3* Pancreatic islets adapt to insulin resistance of pregnancy by up regulating β‑cell mass and increasing insulin secretion. Previously, using a transgenic mouse with global, heterozygous deletion of prolactin receptor (Prlr+/−), we found Prlr signaling is important for this adaptation. However, since Prlr is expressed in tissues outside of islets as well as within islets and prolactin signaling afects β‑cell development, to understand β‑cell‑specifc efect of prolactin signaling in pregnancy, we generated a transgenic mouse with an inducible conditional deletion of Prlr from β‑cells. Here, we found that β‑cell‑specifc Prlr reduction in adult mice led to elevated blood glucose, lowed β‑cell mass and blunted in vivo glucose‑stimulated insulin secretion during pregnancy. When we compared gene expression profle of islets from transgenic mice with global (Prlr+/−) versus β‑cell‑specifc Prlr reduction (βPrlR+/−), we found 95 diferentially expressed gene, most of them down regulated in the Prlr+/− mice in comparison to the βPrlR+/− mice, and many of these genes regulate apoptosis, synaptic vesicle function and neuronal development. Importantly, we found that islets from pregnant Prlr+/− mice are more vulnerable to glucolipotoxicity‑induced apoptosis than islets from pregnant βPrlR+/− mice. These observations suggest that down regulation of prolactin action during pregnancy in non‑β‑cells secondarily and negatively afect β‑cell gene expression, and increased β‑cell susceptibility to external insults. -

Identification of Multiple Risk Loci and Regulatory Mechanisms Influencing Susceptibility to Multiple Myeloma
Corrected: Author correction ARTICLE DOI: 10.1038/s41467-018-04989-w OPEN Identification of multiple risk loci and regulatory mechanisms influencing susceptibility to multiple myeloma Molly Went1, Amit Sud 1, Asta Försti2,3, Britt-Marie Halvarsson4, Niels Weinhold et al.# Genome-wide association studies (GWAS) have transformed our understanding of susceptibility to multiple myeloma (MM), but much of the heritability remains unexplained. 1234567890():,; We report a new GWAS, a meta-analysis with previous GWAS and a replication series, totalling 9974 MM cases and 247,556 controls of European ancestry. Collectively, these data provide evidence for six new MM risk loci, bringing the total number to 23. Integration of information from gene expression, epigenetic profiling and in situ Hi-C data for the 23 risk loci implicate disruption of developmental transcriptional regulators as a basis of MM susceptibility, compatible with altered B-cell differentiation as a key mechanism. Dysregu- lation of autophagy/apoptosis and cell cycle signalling feature as recurrently perturbed pathways. Our findings provide further insight into the biological basis of MM. Correspondence and requests for materials should be addressed to K.H. (email: [email protected]) or to B.N. (email: [email protected]) or to R.S.H. (email: [email protected]). #A full list of authors and their affliations appears at the end of the paper. NATURE COMMUNICATIONS | (2018) 9:3707 | DOI: 10.1038/s41467-018-04989-w | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | DOI: 10.1038/s41467-018-04989-w ultiple myeloma (MM) is a malignancy of plasma cells consistent OR across all GWAS data sets, by genotyping an Mprimarily located within the bone marrow. -

A Silent Exonic SNP in Kdm3a Affects Nucleic Acids Structure but Does Not Regulate Experimental Autoimmune Encephalomyelitis
A Silent Exonic SNP in Kdm3a Affects Nucleic Acids Structure but Does Not Regulate Experimental Autoimmune Encephalomyelitis Alan Gillett1, Petra Bergman1, Roham Parsa1, Andreas Bremges2, Robert Giegerich2, Maja Jagodic1* 1 Department of Clinical Neuroscience, Centre for Molecular Medicine, Karolinska Institutet, Stockholm, Sweden, 2 Center for Biotechnology and Faculty of Technology, Bielefeld University, Bielefeld, Germany Abstract Defining genetic variants that predispose for diseases is an important initiative that can improve biological understanding and focus therapeutic development. Genetic mapping in humans and animal models has defined genomic regions controlling a variety of phenotypes known as quantitative trait loci (QTL). Causative disease determinants, including single nucleotide polymorphisms (SNPs), lie within these regions and can often be identified through effects on gene expression. We previously identified a QTL on rat chromosome 4 regulating macrophage phenotypes and immune-mediated diseases including experimental autoimmune encephalomyelitis (EAE). Gene analysis and a literature search identified lysine-specific demethylase 3A (Kdm3a) as a potential regulator of these phenotypes. Genomic sequencing determined only two synonymous SNPs in Kdm3a. The silent synonymous SNP in exon 15 of Kdm3a caused problems with quantitative PCR detection in the susceptible strain through reduced amplification efficiency due to altered secondary cDNA structure. Shape Probability Shift analysis predicted that the SNP often affects RNA folding; thus, it may impact protein translation. Despite these differences in rats, genetic knockout of Kdm3a in mice resulted in no dramatic effect on immune system development and activation or EAE susceptibility and severity. These results provide support for tools that analyze causative SNPs that impact nucleic acid structures. Citation: Gillett A, Bergman P, Parsa R, Bremges A, Giegerich R, et al. -
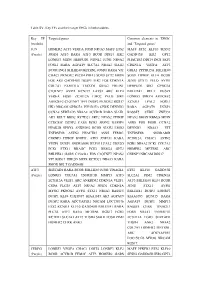
Targeted Genes Common Elements In
Table SV. Key TFs and their target DEGs in hub modules. Key TF Targeted genes Common elements in ‘DEGs’ (module) and ‘Targeted genes’ JUN HOMER2 ATF3 VEGFA FOSB NR4A3 MAFF ETS2 MAFF ETS2 KLF10 SESN2 (Purple) JOSD1 ATF3 RARA ATF3 BCOR DDIT4 IER2 GADD45B IER2 GPT2 LONRF3 MIDN HERPUD1 NDNL2 JUNB NR4A2 PHACTR3 DDIT4 ING1 SKP2 FOSL1 RARA AGPAT9 SLC7A1 NR4A2 SIAH2 CDKN1A VEGFA ATF3 BCOR ING1 BHLHE40 METRNL JOSD1 RARA VIT GRIA2 PPP1R15A BHLHE40 CHAC1 PKNOX2 PLCB4 PHF13 SOX9 SYT2 MIDN SOX9 ITPRIP KLF4 BCOR FOS AK5 GADD45B DUSP1 IER2 FOS CDKN1A JUNB STX11 PELO AVPI1 COL7A1 FAM131A TBCCD1 GRIA2 ERLIN1 HERPUD1 SIK1 GPRC5A C1QTNF7 AVPI1 KCNJ15 LATS2 ARC KLF4 BHLHE41 RELT DUSP2 VEGFA FOSB ZC3H12A LMO2 PELO SIK1 LONRF3 SPRY4 ARHGEF2 ARHGEF2 C1QTNF7 TFPI DUSP2 PKNOX2 RGS17 KCNJ15 LPAL2 FOSL1 HK1 NRCAM GPRC5A PPP1R15A CERK DENND3 RARA AGPAT9 DUSP1 CCNA2 SERTAD3 NR4A2 ACVR1B RARA SIAH1 RASSF5 CERK ZNF331 AK5 RELT BRD2 KCTD21 SKP2 NPAS2 ITPRIP NPAS2 SMOX RBM24 MIDN CCDC85C DUSP2 CARS EGR1 SESN2 RASSF5 ASNS FOS FOSB CCNA2 PIP4K2B SPRY4 ANKRD52 BCOR SIAH2 LMO2 DENND3 NR4A3 VIT TNFRSF1B ASTN2 PHACTR3 ASNS FEM1C TNFRSF1B SNORA80B CSRNP3 ITPRIP BNIP3L ATF3 ZNF331 RARA ZC3H12A CHAC1 ASTN2 VEZF1 DUSP1 SNORA80B KLF10 LPAL2 UBE2O EGR1 NR4A2 PCK1 COL7A1 PCK1 STX11 RRAGC PCK1 RBM24 GPT2 HOMER2 METRNL ARC BHLHE41 GARS C15orf41 FOS C1QTNF7 NPAS2 CSRNP3 NRCAM RGS17 VIT RGS17 UBE2O SOX9 KCTD21 NR4A3 RARA SMOX RELT GADD45B ATF3 SERTAD1 RARA BCOR BHLHE40 JUNB TINAGL1 ETS2 KLF10 GADD45B (Purple) LONRF3 COL7A1 TNFRSF1B MMP13 ATF3 SLC2A1 PIM2 CDKN1A ZC3H12A VEZF1 ARC ANKRD52 -

Downregulation of Mir-335 Exhibited an Oncogenic Effect Via Promoting KDM3A/YAP1 Networks in Clear Cell Renal Cell Carcinoma
Cancer Gene Therapy https://doi.org/10.1038/s41417-021-00335-3 ARTICLE Downregulation of miR-335 exhibited an oncogenic effect via promoting KDM3A/YAP1 networks in clear cell renal cell carcinoma 1 1 1 1 1 1 1 Wenqiang Zhang ● Ruiyu Liu ● Lin Zhang ● Chao Wang ● Ziyan Dong ● Jiasheng Feng ● Mayao Luo ● 1 1 1 1 Yifan Zhang ● Zhuofan Xu ● Shidong Lv ● Qiang Wei Received: 23 December 2020 / Revised: 3 March 2021 / Accepted: 26 March 2021 © The Author(s) 2021. This article is published with open access Abstract Clear cell renal cell carcinoma (ccRCC) is the most common type of renal cancer affecting many people worldwide. Although the 5-year survival rate is 65% in localized disease, after metastasis, the survival rate is <10%. Emerging evidence has shown that microRNAs (miRNAs) play a crucial regulatory role in the progression of ccRCC. Here, we show that miR- 335, an anti-onco-miRNA, is downregulation in tumor tissue and inhibited ccRCC cell proliferation, invasion, and migration. Our studies further identify the H3K9me1/2 histone demethylase KDM3A as a new miR-335-regulated gene. We show that KDM3A is overexpressed in ccRCC, and its upregulation contributes to the carcinogenesis and metastasis of fi 1234567890();,: 1234567890();,: ccRCC. Moreover, with the overexpression of KDM3A, YAP1 was increased and identi ed as a direct downstream target of KDM3A. Enrichment of KDM3A demethylase on YAP1 promoter was confirmed by CHIP-qPCR and YAP1 was also found involved in the cell growth and metastasis inhibitory of miR-335. Together, our study establishes a new miR-335/ KDM3A/YAP1 regulation axis, which provided new insight and potential targeting of the metastasized ccRCC. -

Histone Methylation Regulation in Neurodegenerative Disorders
International Journal of Molecular Sciences Review Histone Methylation Regulation in Neurodegenerative Disorders Balapal S. Basavarajappa 1,2,3,4,* and Shivakumar Subbanna 1 1 Division of Analytical Psychopharmacology, Nathan Kline Institute for Psychiatric Research, Orangeburg, NY 10962, USA; [email protected] 2 New York State Psychiatric Institute, New York, NY 10032, USA 3 Department of Psychiatry, College of Physicians & Surgeons, Columbia University, New York, NY 10032, USA 4 New York University Langone Medical Center, Department of Psychiatry, New York, NY 10016, USA * Correspondence: [email protected]; Tel.: +1-845-398-3234; Fax: +1-845-398-5451 Abstract: Advances achieved with molecular biology and genomics technologies have permitted investigators to discover epigenetic mechanisms, such as DNA methylation and histone posttransla- tional modifications, which are critical for gene expression in almost all tissues and in brain health and disease. These advances have influenced much interest in understanding the dysregulation of epigenetic mechanisms in neurodegenerative disorders. Although these disorders diverge in their fundamental causes and pathophysiology, several involve the dysregulation of histone methylation- mediated gene expression. Interestingly, epigenetic remodeling via histone methylation in specific brain regions has been suggested to play a critical function in the neurobiology of psychiatric disor- ders, including that related to neurodegenerative diseases. Prominently, epigenetic dysregulation currently brings considerable interest as an essential player in neurodegenerative disorders, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), Amyotrophic lateral sclerosis (ALS) and drugs of abuse, including alcohol abuse disorder, where it may facilitate connections between genetic and environmental risk factors or directly influence disease-specific Citation: Basavarajappa, B.S.; Subbanna, S. -

The Emerging Role of Histone Lysine Demethylases in Prostate Cancer
Crea et al. Molecular Cancer 2012, 11:52 http://www.molecular-cancer.com/content/11/1/52 REVIEW Open Access The emerging role of histone lysine demethylases in prostate cancer Francesco Crea1*, Lei Sun3, Antonello Mai4, Yan Ting Chiang1, William L Farrar3, Romano Danesi2 and Cheryl D Helgason1,5* Abstract Early prostate cancer (PCa) is generally treatable and associated with good prognosis. After a variable time, PCa evolves into a highly metastatic and treatment-refractory disease: castration-resistant PCa (CRPC). Currently, few prognostic factors are available to predict the emergence of CRPC, and no curative option is available. Epigenetic gene regulation has been shown to trigger PCa metastasis and androgen-independence. Most epigenetic studies have focused on DNA and histone methyltransferases. While DNA methylation leads to gene silencing, histone methylation can trigger gene activation or inactivation, depending on the target amino acid residues and the extent of methylation (me1, me2, or me3). Interestingly, some histone modifiers are essential for PCa tumor- initiating cell (TIC) self-renewal. TICs are considered the seeds responsible for metastatic spreading and androgen- independence. Histone Lysine Demethylases (KDMs) are a novel class of epigenetic enzymes which can remove both repressive and activating histone marks. KDMs are currently grouped into 7 major classes, each one targeting a specific methylation site. Since their discovery, KDM expression has been found to be deregulated in several neoplasms. In PCa, KDMs may act as either tumor suppressors or oncogenes, depending on their gene regulatory function. For example, KDM1A and KDM4C are essential for PCa androgen-dependent proliferation, while PHF8 is involved in PCa migration and invasion.