Mast Cells in Asthma: Biomarker and Therapeutic Target
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Peripheral Regulation of Pain and Itch
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 1596 Peripheral Regulation of Pain and Itch ELÍN INGIBJÖRG MAGNÚSDÓTTIR ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6206 ISBN 978-91-513-0746-6 UPPSALA urn:nbn:se:uu:diva-392709 2019 Dissertation presented at Uppsala University to be publicly examined in A1:107a, BMC, Husargatan 3, Uppsala, Friday, 25 October 2019 at 13:00 for the degree of Doctor of Philosophy (Faculty of Medicine). The examination will be conducted in English. Faculty examiner: Professor emeritus George H. Caughey (University of California, San Francisco). Abstract Magnúsdóttir, E. I. 2019. Peripheral Regulation of Pain and Itch. Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 1596. 71 pp. Uppsala: Acta Universitatis Upsaliensis. ISBN 978-91-513-0746-6. Pain and itch are diverse sensory modalities, transmitted by the somatosensory nervous system. Stimuli such as heat, cold, mechanical pain and itch can be transmitted by different neuronal populations, which show considerable overlap with regards to sensory activation. Moreover, the immune and nervous systems can be involved in extensive crosstalk in the periphery when reacting to these stimuli. With recent advances in genetic engineering, we now have the possibility to study the contribution of distinct neuron types, neurotransmitters and other mediators in vivo by using gene knock-out mice. The neuropeptide calcitonin gene-related peptide (CGRP) and the ion channel transient receptor potential cation channel subfamily V member 1 (TRPV1) have both been implicated in pain and itch transmission. In Paper I, the Cre- LoxP system was used to specifically remove CGRPα from the primary afferent population that expresses TRPV1. -

The Emerging Role of Mast Cell Proteases in Asthma
REVIEW ASTHMA The emerging role of mast cell proteases in asthma Gunnar Pejler1,2 Affiliations: 1Dept of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden. 2Dept of Anatomy, Physiology and Biochemistry, Swedish University of Agricultural Sciences, Uppsala, Sweden. Correspondence: Gunnar Pejler, Dept of Medical Biochemistry and Microbiology, BMC, Uppsala University, Box 582, 75123 Uppsala, Sweden. E-mail: [email protected] @ERSpublications Mast cells express large amounts of proteases, including tryptase, chymase and carboxypeptidase A3. An extensive review of how these proteases impact on asthma shows that they can have both protective and detrimental functions. http://bit.ly/2Gu1Qp2 Cite this article as: Pejler G. The emerging role of mast cell proteases in asthma. Eur Respir J 2019; 54: 1900685 [https://doi.org/10.1183/13993003.00685-2019]. ABSTRACT It is now well established that mast cells (MCs) play a crucial role in asthma. This is supported by multiple lines of evidence, including both clinical studies and studies on MC-deficient mice. However, there is still only limited knowledge of the exact effector mechanism(s) by which MCs influence asthma pathology. MCs contain large amounts of secretory granules, which are filled with a variety of bioactive compounds including histamine, cytokines, lysosomal hydrolases, serglycin proteoglycans and a number of MC-restricted proteases. When MCs are activated, e.g. in response to IgE receptor cross- linking, the contents of their granules are released to the exterior and can cause a massive inflammatory reaction. The MC-restricted proteases include tryptases, chymases and carboxypeptidase A3, and these are expressed and stored at remarkably high levels. -

Download PDF File
CLINICAL CARDIOLOGY Cardiology Journal 2019, Vol. 26, No. 6, 680–686 DOI: 10.5603/CJ.a2018.0018 Copyright © 2019 Via Medica ORIGINAL ARTICLE ISSN 1897–5593 Mast cell derived carboxypeptidase A3 is decreased among patients with advanced coronary artery disease Łukasz Lewicki1, 2, 3, Janusz Siebert1, 4, Tomasz Koliński5, Karolina Piekarska5, Magdalena Reiwer-Gostomska4, Radosław Targoński3, Piotr Trzonkowski6, Natalia Marek-Trzonkowska5 1University Center for Cardiology, Gdansk, Poland 2Department of Machine Design and Automotive Engineering, Faculty of Mechanical Engineering, Gdansk University of Technology, Gdansk, Poland 3Pomeranian Cardiology Centers, Wejherowo, Poland 4Department of Family Medicine, Medical University of Gdansk, Poland 6Department of Family Medicine, Laboratory of Immunoregulation and Cellular Therapies, Medical University of Gdansk, Poland 7Department of Clinical Immunology and Transplantology, Medical University of Gdansk, Poland Abstract Background: Coronary artery disease (CAD) affects milions of people and can result in myocardial infarction (MI). Previously, mast cells (MC) have been extensively investigated in the context of hyper- sensitivity, however as regulators of the local inflammatory response they can potentially contribute to CAD and/or its progression. The aim of the study was to assess if serum concentration of MC proteases: carboxypeptidase A3, cathepsin G and chymase 1 is associated with the extension of CAD and MI. Methods: The 44 patients with angiographically confirmed CAD (23 subjects with non-ST-segment elevation MI [NSTEMI] and 21 with stable CAD) were analyzed. Clinical data were obtained as well serum concentrations of carboxypeptidase A3, cathepsin G and chymase 1 were also measured. Results: Patients with single vessel CAD had higher serum concentration of carboxypeptidase than those with more advanced CAD (3838.6 ± 1083.1 pg/mL vs. -

Extracellular Matrix Proteomics Identifies Molecular Signature of Symptomatic Carotid Plaques
CLINICAL MEDICINE The Journal of Clinical Investigation Extracellular matrix proteomics identifies molecular signature of symptomatic carotid plaques Sarah R. Langley,1,2 Karin Willeit,3 Athanasios Didangelos,1 Ljubica Perisic Matic,4 Philipp Skroblin,1 Javier Barallobre-Barreiro,1 Mariette Lengquist,4 Gregor Rungger,5 Alexander Kapustin,1 Ludmilla Kedenko,6 Chris Molenaar,1,7 Ruifang Lu,1 Temo Barwari,1 Gonca Suna,1 Xiaoke Yin,1 Bernhard Iglseder,8 Bernhard Paulweber,6 Peter Willeit,3,9 Joseph Shalhoub,10 Gerard Pasterkamp,11 Alun H. Davies,10 Claudia Monaco,12 Ulf Hedin,4 Catherine M. Shanahan,1 Johann Willeit,3 Stefan Kiechl,3 and Manuel Mayr1 1King’s British Heart Foundation Centre, King’s College London, London, United Kingdom. 2Duke-NUS Medical School, Singapore. 3Department of Neurology, Medical University Innsbruck, Innsbruck, Austria. 4Department of Molecular Medicine and Surgery, Vascular Surgery, Karolinska Institute, Stockholm, Sweden. 5Department of Neurology, Bruneck Hospital, Bruneck, Italy. 6First Department of Internal Medicine, Paracelsus Medical University, Salzburg, Austria. 7Nikon Imaging Centre, King’s College London, London, United Kingdom. 8Department of Geriatric Medicine, Paracelsus Medical University, Salzburg, Austria. 9Department of Public Health and Primary Care, University of Cambridge, Cambridge, United Kingdom. 10Department of Surgery and Cancer, Imperial College London, London, United Kingdom. 11Laboratory of Clinical Chemistry and Experimental Cardiology, University Medical Center Utrecht, Utrecht, The Netherlands. 12Kennedy Institute, University of Oxford, Oxford, United Kingdom BACKGROUND. The identification of patients with high-risk atherosclerotic plaques prior to the manifestation of clinical events remains challenging. Recent findings question histology- and imaging-based definitions of the “vulnerable plaque,” necessitating an improved approach for predicting onset of symptoms. -

The Imprinted Region on Human Chromosome 7Q32 Extends to The
249 ORIGINAL ARTICLE J Med Genet: first published as 10.1136/jmg.40.4.249 on 1 April 2003. Downloaded from The imprinted region on human chromosome 7q32 extends to the carboxypeptidase A gene cluster: an imprinted candidate for Silver-Russell syndrome L Bentley, K Nakabayashi, D Monk, C Beechey, J Peters, Z Birjandi, F E Khayat, M Patel, M A Preece, P Stanier, S W Scherer, G E Moore ............................................................................................................................. J Med Genet 2003;40:249–256 See end of article for authors’ affiliations Imprinted gene(s) on human chromosome 7q32-qter have been postulated to be involved in intrauterine ....................... growth restriction associated with Silver-Russell syndrome (SRS) as 7-10% of patients have mUPD(7). Correspondence to: Three imprinted genes, MEST, MESTIT1, and COPG2IT1 on chromosome 7q32, are unlikely to cause L Bentley, Department of SRS since epigenetic and sequence mutation analyses have not shown any changes. One hundred Fetal and Maternal kilobases proximal to MEST lies a group of four carboxypeptidase A (CPA) genes. Since most imprinted Medicine, Institute of genes are found in clusters, this study focuses on analysing these CPAs for imprinting effects based on Reproductive and Developmental Biology, their proximity to an established imprinted domain. Firstly, a replication timing study across 7q32 Faculty of Medicine, showed that an extensive genomic region including the CPAs, MEST, MESTIT1, and COPG2IT1 repli- Imperial College, cates asynchronously. Subsequently, SNP analysis by sequencing RT-PCR products of CPA1, CPA2, Hammersmith Campus, CPA4, and CPA5 indicated preferential expression of CPA4. Pyrosequencing was used as a quantita- Du Cane Road, London tive approach, which confirmed predominantly preferential expression of the maternal allele and bial- W12 0NN, UK; [email protected] lelic expression in brain. -

Molecular Mechanism of Mast Cell–Mediated Innate Defense Against Endothelin and Snake Venom Sarafotoxin
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/5920550 Molecular mechanism of mast cell–mediated innate defense against endothelin and snake venom sarafotoxin Article in Journal of Experimental Medicine · November 2007 DOI: 10.1084/jem.20071262 · Source: PubMed CITATIONS READS 89 75 5 authors, including: Lars Alexander Schneider Susan Mariola Schlenner Ulm University Vlaams Instituut voor Biotechnologie and KU L… 101 PUBLICATIONS 1,445 CITATIONS 28 PUBLICATIONS 898 CITATIONS SEE PROFILE SEE PROFILE Markus Wunderlin Ulm University 14 PUBLICATIONS 473 CITATIONS SEE PROFILE All content following this page was uploaded by Susan Mariola Schlenner on 01 June 2014. The user has requested enhancement of the downloaded file. ARTICLE Molecular mechanism of mast cell– mediated innate defense against endothelin and snake venom sarafotoxin Lars A. Schneider, 1 Susan M. Schlenner,1 Thorsten B. Feyerabend, 1 Markus Wunderlin,2 and Hans-Reimer Rodewald 1 1 Institute for Immunology, 2 Section for Mass Spectrometry, Institute for Organic Chemistry II, University of Ulm, D-89081 Ulm, Germany Mast cells are protective against snake venom sarafotoxins that belong to the endothelin (ET) peptide family. The molecular mechanism underlying this recently recognized innate defense pathway is unknown, but secretory granule proteases have been invoked. To spe- cifi cally disrupt a single protease function without affecting expression of other proteases, we have generated a mouse mutant selectively lacking mast cell carboxypeptidase A (Mc-cpa) activity. Using this mutant, we have now identifi ed Mc-cpa as the essential protective mast cell enzyme. Mass spectrometry of peptide substrates after cleavage by normal or mutant mast cells showed that removal of a single amino acid, the C-terminal tryptophan, from ET and sarafotoxin by Mc-cpa is the principle molecular mechanism underlying this very rapid mast cell response. -

Mast Cells with Multiple Protease Deficiency Distorted Secretory
Distorted Secretory Granule Composition in Mast Cells with Multiple Protease Deficiency Mirjana Grujic, Gabriela Calounova, Inger Eriksson, Thorsten Feyerabend, Hans-Reimer Rodewald, Elena This information is current as Tchougounova, Lena Kjellén and Gunnar Pejler of October 1, 2021. J Immunol 2013; 191:3931-3938; Prepublished online 23 August 2013; doi: 10.4049/jimmunol.1301441 http://www.jimmunol.org/content/191/7/3931 Downloaded from References This article cites 29 articles, 16 of which you can access for free at: http://www.jimmunol.org/content/191/7/3931.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on October 1, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Distorted Secretory Granule Composition in Mast Cells with Multiple Protease Deficiency Mirjana Grujic,* Gabriela Calounova,*,1 Inger Eriksson,†,1 Thorsten Feyerabend,‡ Hans-Reimer Rodewald,‡ Elena Tchougounova,x Lena Kjelle´n,† and Gunnar Pejler* Mast cells are characterized by an abundance of secretory granules densely packed with inflammatory mediators such as bioactive amines, cytokines, serglycin proteoglycans with negatively charged glycosaminoglycan side chains of either heparin or chondroitin sulfate type, and large amounts of positively charged proteases. -
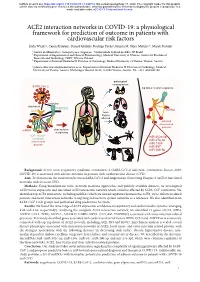
ACE2 Interaction Networks in COVID-19
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.13.094714; this version posted May 17, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors Zofia Wicik1,2, Ceren Eyileten2, Daniel Jakubik2, Rodrigo Pavão1, Jolanta M. Siller-Matula2,3*, Marek Postula2 ¹ Centro de Matemática, Computação e Cognição - Universidade Federal do ABC, SP, Brazil ² Department of Experimental and Clinical Pharmacology, Medical University of Warsaw, Center for Preclinical Research and Technology CEPT, Warsaw, Poland ³ Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Vienna, Austria * [email protected]; Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Austria. Waehringer Guertel 18-20, A-1090 Vienna, Austria. Tel. +43 1 4040046140 pathological consequences SARS-CoV-2 top ACE2 network regulators activation attachment TMPRSS2 miRNA ACE2 main tissues affected activated virus-related proteins miRNA dysregulation of signaling ACE2 network miR-302c-5p containing miR-27a-3p virus-related proteins miR-1305 miR-587 miR-26b-5p Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (coronavirus disease 2019; COVID-19) is associated with adverse outcome in patients with cardiovascular disease (CVD). Aim: To characterize the interaction between SARS-CoV-2 and Angiotensin Converting Enzyme 2 (ACE2) functional networks with focus on CVD. -

Regulation of Early T-Lineage Gene Expression and Developmental Progression by the Progenitor Cell Transcription Factor PU.1
Downloaded from genesdev.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1 Ameya Champhekar,1,4 Sagar S. Damle,1,5 George Freedman,1,6 Sebastian Carotta,2,3 Stephen L. Nutt,2,3 and Ellen V. Rothenberg1 1Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, California 91125, USA; 2Division of Molecular Immunology, Walter and Eliza Hall Institute of Medical Research, Melbourne, Victoria 3052, Australia; 3Department of Medical Biology, The University of Melbourne, Parkville, Victoria 3010, Australia The ETS family transcription factor PU.1 is essential for the development of several blood lineages, including T cells, but its function in intrathymic T-cell precursors has been poorly defined. In the thymus, high PU.1 expression persists through multiple cell divisions in early stages but then falls sharply during T-cell lineage commitment. PU.1 silencing is critical for T-cell commitment, but it has remained unknown how PU.1 activities could contribute positively to T-cell development. Here we employed conditional knockout and modified antagonist PU.1 constructs to perturb PU.1 function stage-specifically in early T cells. We show that PU.1 is needed for full proliferation, restricting access to some non-T fates, and controlling the timing of T-cell developmental progression such that removal or antagonism of endogenous PU.1 allows precocious access to T-cell differentiation. Dominant-negative effects reveal that this repression by PU.1 is mediated indirectly. Genome-wide transcriptome analysis identifies novel targets of PU.1 positive and negative regulation affecting progenitor cell signaling and cell biology and indi- cating distinct regulatory effects on different subsets of progenitor cell transcription factors. -

An Update on the Tissue Renin Angiotensin System and Its Role in Physiology and Pathology
Journal of Cardiovascular Development and Disease Review An Update on the Tissue Renin Angiotensin System and Its Role in Physiology and Pathology Ali Nehme 1 , Fouad A. Zouein 2 , Zeinab Deris Zayeri 3 and Kazem Zibara 4,* 1 EA4173, Functional genomics of arterial hypertension, Univeristy Claude Bernard Lyon-1 (UCBL-1), 69008 Lyon, France; [email protected] 2 Department of Pharmacology and Toxicology, Heart Repair Division, Faculty of Medicine, American University of Beirut, Beirut 11-0236, Lebanon; [email protected] 3 Thalassemia & Hemoglobinopathy Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; [email protected] 4 PRASE, Biology Department, Faculty of Sciences-I, Lebanese University, Beirut, Lebanon * Correspondence: [email protected] Received: 10 February 2019; Accepted: 26 March 2019; Published: 29 March 2019 Abstract: In its classical view, the renin angiotensin system (RAS) was defined as an endocrine system involved in blood pressure regulation and body electrolyte balance. However, the emerging concept of tissue RAS, along with the discovery of new RAS components, increased the physiological and clinical relevance of the system. Indeed, RAS has been shown to be expressed in various tissues where alterations in its expression were shown to be involved in multiple diseases including atherosclerosis, cardiac hypertrophy, type 2 diabetes (T2D) and renal fibrosis. In this chapter, we describe the new components of RAS, their tissue-specific expression, and their alterations under pathological conditions, which will help achieve more tissue- and condition-specific treatments. Keywords: renin-angiotensin-aldosterone system; tissue; expression; physiology 1. Introduction In its classical view, RAS was defined as an endocrine system involved in blood pressure regulation and body electrolyte balance. -

Protease Phenotype of Constitutive Connective Tissue and of Induced Mucosal Mast Cells in Mice Is Regulated by the Tissue
Protease phenotype of constitutive connective tissue and of induced mucosal mast cells in mice is regulated by the tissue Wei Xing, K. Frank Austen1, Michael F. Gurish1, and Tatiana G. Jones Division of Rheumatology, Immunology and Allergy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA 02115 Contributed by K. Frank Austen, July 11, 2011 (sent for review June 24, 2011) Mouse mast cells (MCs) express a large number of serine proteases The human MC expresses a tryptase, beta 1, a chymase, and including tryptases, mouse mast cell protease (mMCP)-6 and -7; a carboxypeptidase that are homologous to mMCP-6, mMCP-4, chymases, mMCP-1, -2, and -4; and an elastase, mMCP-5; along and mouse CPA3, respectively (10, 14). These proteases are with carboxypeptidase-A3 (CPA3). In helminth-infected mouse in- expressed in intestinal and skin CTMC (often termed MCTC), but testine, distinct protease phenotypes are observed for connective only tryptase is present in most MMCs (termed MCT) of intestine tissue MCs (CTMCs) (mMCP-4+–7+, and CPA3+) and mucosal MCs and lung (15, 16). The MMCs are considered to be T-cell (MMCs) (mMCP-1+ and 2+). To determine whether the protease dependent, on the basis of their absence in the gastrointestinal phenotype was regulated by the tissue, we compared the pheno- mucosa of individuals with acquired severe immune deficiency type of constitutive CTMCs and induced MMCs in trachea and large (17). Recently, the reported protease exclusivity of these two airways in antigen-sensitized unchallenged and challenged mice phenotypes in human MCs has been challenged by the findings of to MCs in skin and helminthic-infected intestine. -

Mammalian Metallopeptidase Inhibition at the Defense Barrier of Ascaris Parasite
Mammalian metallopeptidase inhibition at the defense barrier of Ascaris parasite Laura Sanglasa, Francesc X. Avilesa, Robert Huberb,c,d,1, F. Xavier Gomis-Ru¨ the,1, and Joan L. Arolasa,e,1 aInstitut de Biotecnologia i de Biomedicina and Departament de Bioquímica i Biologia Molecular, Facultat de Cie`ncies, Universitat Auto`noma de Barcelona, E-08193 Bellaterra, Barcelona, Spain; bMax-Planck-Institut fu¨r Biochemie, Am Klopferspitz 18a, D-82152 Martinsried, Germany; cSchool of Biosciences, Cardiff University, Cardiff CF10 3US, United Kingdom; dCentre for Medical Biotechnology, Fachbereich Biology and Geography, University Duisburg-Essen, Universita¨tsstrasse, D-45117 Essen, Germany; and eDepartment of Structural Biology, Proteolysis Laboratory, Molecular Biology Institute of Barcelona, Consejo Superior de Investigaciones Cientificas, Barcelona Science Park, Helix Building, c/Baldiri Reixac 15-21, E-08028 Barcelona, Spain Contributed by Robert Huber, December 12, 2008 (sent for review November 17, 2008) Roundworms of the genus Ascaris are common parasites of the catalyze the hydrolysis of C-terminal amino acids from proteins human gastrointestinal tract. A battery of selective inhibitors and peptides. They perform a large variety of physiologically protects them from host enzymes and the immune system. Here, a relevant functions in organisms of different phyla (17). These metallocarboxypeptidase (MCP) inhibitor, ACI, was identified in enzymes have been grouped into the funnelin tribe of proteases protein extracts from Ascaris by intensity-fading MALDI-TOF mass and are subdivided into A/B- and N/E-type MCPs (18). Human spectrometry. The 67-residue amino acid sequence of ACI showed A/B-type funnelins include the digestive enzymes CPA1, CPA2, no significant homology with any known protein.