Molecular Diversity in Earias Insulana Populations from Different Egyptian Governorates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Opportunities and Challenges Faced with Emerging Technologies in the Australian Vegetable Industry
HAL Project Title: Opportunities and challenges faced with emerging technologies in the Australian vegetable industry. (Technology Platform 5: Emerging Technologies for Production and Harvest) Project VG08087 Project completion date: 30/06/2010 Author: Dr Silvia Estrada-Flores VG08087 Emerging Technologies: Production Opportunities and challenges faced with emerging technologies in the Australian vegetable industry. (Technology Platform 5: Emerging Technologies for Production and Harvest) Horticulture Australia Project Number: VG08087 Project Leader: Dr Silvia Estrada-Flores. Principal Consultant, Food Chain Intelligence. Contact details: PO Box 1789. North Sydney 2059, NSW. Ph 0404 353 571; e-mail: [email protected] &RPSDQ\¶VZebsite: www.food-chain.com.au Delivered on: June 2010. Purpose of Project: This report was prepared as an outcome of Milestone 190 RISURMHFW9*´2SSRUWXQLWLHVDQG challenJHVIDFHGZLWKHPHUJLQJWHFKQRORJLHVLQWKH$XVWUDOLDQYHJHWDEOHLQGXVWU\´7KHSURMHFW aims to provide a broad review of technologies that are influencing the competitiveness of the industry. This is the fourth of five reports to be developed during 2009-2010 and reviews novel production and harvesting technologies for vegetables. Funding acknowledgements: Food Chain Intelligence acknowledges the financial support for this project from Horticulture Australia Limited (HAL) and AUSVEG. Disclaimers: Any recommendations contained in this publication do not necessarily represent current HAL policy. No person should act on the basis of the contents of this publication, -

And Earias Vittella (Lepidoptera: Noctuidae)
IOSR Journal of Pharmacy and Biological Sciences (IOSR-JPBS) ISSN: 2278-3008. Volume 4, Issue 6 (Jan. – Feb. 2013), PP 09-12 www.iosrjournals.org Pesticidal activity of Pakistani Bacillus thuringiensis isolates against Helicoverpa armigera (Hubner) and Earias vittella (Lepidoptera: Noctuidae). 1Kausar Malik, 2Farkhanda Jabeen, 3Mir Muhammad Ali Talpur, 4Shagufta Andleeb and 5Amjad Farooq 1Department of Zoology, Lahore College for Women University, Lahore, Pakistan. 2,3Former address: National Centre of Excellence in Molecular Biology, University of the Punjab, Lahore, Pakistan. Abstract: A large number of Bacillus thuringiensis isolates separated from different ecological regions of Pakistan were analysed for pesticidal activity against two lepidopteran cotton insect pests, American bollworm (Helicoverpa armigera and spotted bollworm (Earias vittella). The biological activity of local B.t. isolates demonstrated a wide range of LC50 values against both target pests. The most potent isolates HW 4.4 and INS2.25 against Helicoverpa armigera showed LC50 value of 9ng/mg of artificial diet. The Lc50 value of 2ng/mg of artificial diet was exhibited by local B.t. isolates HFZ 11.3, MR 19.1 and MG 2.6 against Earias vittella. Keywords:Pakistani, Bacillus thuringiensis, Earias vittella, Helicoverpa armigera, Isolate, Lepidoptera, Noctuidae, I. Introduction Microbial control of insect pest of crops using entomopathogens is an ecologically sound pest management strategy. Although insect viruses and fungal pathogens are used as microbial control agents, but Bacillus thuringiensis Berliner (B.t.) appears to has the greatest potential for this purpose. Bacillus thuringiensis is an aerobic, spore forming, Gram-positive bacterium that synthesizes a crystalline parasporal inclusion composed of one of several proteins known as insecticidal crystal proteins (ICP) or delta-endotoxins during sporulation. -

Assessment of Some Botanicals on the Life-Table
Pratibha & Singh RJLBPCS 2018 www.rjlbpcs.com Life Science Informatics Publications Original Research Article DOI: 10.26479/2018.0406.53 ASSESSMENT OF SOME BOTANICALS ON THE LIFE-TABLE PARAMETERS OF OKRA MOTH, EARIAS VITTELLA (FABR.) (LEPIDOPTERA: NOCTUIDAE) Pratibha, Rajendra Singh* Department of Zoology, D.D.U. Gorakhpur University, Gorakhpur, U. P, India ABSTRACT: The okra moth Earias vittella is one of the dangerous pests attacking okra in India. The present study was conducted aiming at evaluation of effects of various botanicals, viz., aqueous extract of garlic bulb (AEG), aqueous extract of neem leaves (AEN) and NeemGold (NG) on the life-table parameters of this insect. Different concentrations of each botanical had been applied on the host plant, Abelmoschus esculentus. The current investigation recorded significant reduction of the age-specific survivorship, reproductive and net fecundity rates; intrinsic rate of natural increase and doubling time of the population of the pest while generation time remains unaffected. Also, longevity of the adult females was remarkably shortened. The strongest shortening action was determined by NG, followed by AEG and AEN, respectively. Total fecundity rate (Rt) was reduced to 102.17 (AEG), 68.00 (AEN), 54.67 (NG), 235.67 progeny/female (Control), respectively and net reproductive rate (Ro) were 52.00 (AEG), 33.83 (AEN), 26.50 (NG) and control was 119.67daughter/female. Likewise, values of the intrinsic rate (rm) were 0.1689 (AEG), 0.1524 (AEN), 0.1440 (NG), 0.2035 (control), respectively. The generation time (GT) insignificantly varied between 22.79 to23.51 days. Reproductive periods and post-reproductive periods were significantly affected (decreased) by the tested botanicals. -

(Sfna) Insecticidal and Inhibitory Potential of Two
Sustainability in Food and Agriculture (SFNA)2(2) (2021) 69-73 Sustainability in Food and Agriculture (SFNA) DOI: http://doi.org/10.26480/sfna.02.2021.69.73 ISSN: 2716-6716 (Online) CODEN: SFAUBO RESEARCH ARTICLE INSECTICIDAL AND INHIBITORY POTENTIAL OF TWO CHITIN SYNTHESIS INHIBITORS, BUPROFEZIN AND LUFENURON ON OKRA SHOOT AND FRUIT BORER, EARIAS VITTELLA Asfia Sharmin, Gopal Das* Department of Entomology, Bangladesh Agricultural University, Mymensingh-2202, Bangladesh. *Corresponding Author Email: [email protected] This is an open access article distributed under the Creative Commons Attribution License CC BY 4.0, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ARTICLE DETAILS ABSTRACT Article History: In this laboratory study, insecticidal and inhibitory potential of two chitin synthesis inhibitors (CSIs) viz. Buprofezin and Lufenuron were evaluated against okra shoot and fruit borer (OSFB). CSIs were applied on Received 01 March 2021 three days old larvae through different bioassay methods like topical or direct, okra-dip or indirect and Accepted 06 April 2021 Available online April 2021 combined (topical + leaf-dip). Data were collected on larval mortality, weight reduction and deformations of larvae and adults. Results showed that larval mortality and weight reduction were clearly dose, application 22 methods and time dependent. In case of both CSIs, the highest mortality was found @ 1.0 ml/L that was followed by 0.75 ml and 0.50 ml/L respectively. Likewise, the highest body weight reduction was also found from 1.0 ml/L. In both cases, the concentration 0.5 ml/L was found less effective. -
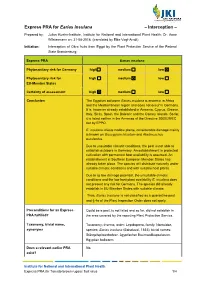
Earias Insulana – Interception –
Express PRA for Earias insulana – Interception – Prepared by: Julius Kuehn-Institute, Institute for National and International Plant Health; Dr. Anne Wilstermann on: 31-08-2018. (translated by Elke Vogt-Arndt) Initiation: Interception of Okra fruits from Egypt by the Plant Protection Service of the Federal State Brandenburg Express-PRA Earias insulana Phytosanitary risk for Germany high medium low Phytosanitary risk for high medium low EU-Member States Certainty of assessment high medium low Conclusion The Egyptian bollworm Earias insulana is endemic in Africa and the Mediterranean region and does not occur in Germany. It is, however already established in Armenia, Cyprus, Greece, Italy, Sicily, Spain, the Balearic and the Canary Islands. So far, it is listed neither in the Annexes of the Directive 2000/29/EC nor by EPPO. E. insulana infests mallow plants, considerable damage mainly is known on Gossypium hirsutum and Abelmoschus esculentus. Due to unsuitable climatic conditions, the pest is not able to establish outdoors in Germany. An establishment in protected cultivation with permanent host availability is assumed. An establishment in Southern European Member States has already taken place. The species will distribute naturally under suitable climatic conditions and with suitable host plants. Due to its low damage potential, the unsuitable climatic conditions and the low host plant availability E. insulana does not present any risk for Germany. The species did already establish in EU-Member States with suitable climate. Thus, Earias insulana is not classified as a quarantine pest and § 4a of the Plant Inspection Order does not apply. Preconditions for an Express- Could be a pest, is not listed and so far, did not establish in PRA fulfilled? the area covered by the reporting Plant Protection Service. -

Ent18 2 117 121 (Kravchenko Et Al).Pmd
Russian Entomol. J. 18(2): 117121 © RUSSIAN ENTOMOLOGICAL JOURNAL, 2009 The Eariadinae and Chloephorinae (Lepidoptera: Noctuoidea, Nolidae) of Israel: distribution, phenology and ecology Eariadinae è Chloephorinae (Lepidoptera: Noctuoidea, Nolidae) Èçðàèëÿ: ðàñïðåäåëåíèå, ôåíîëîãèÿ è ýêîëîãèÿ V.D. Kravchenko1, Th. Witt2, W. Speidel2, J. Mooser3, A. Junnila4 & G.C. Müller4 Â.Ä. Êðàâ÷åíêî1,Ò. Âèòò2, Â. Øïàéäåëü2, Äæ. Ìîçåð3, Ý. Äæàííèëà4 , Ã.Ê. Ìþëëåð4 1 Department of Zoology, Tel Aviv University, Tel Aviv, Israel. 2 Museum Witt, Tengstr. 33, D-80796 Munich, Germany. 3 Seilerbruecklstr. 23, D-85354 Freising, Germany. 4 Department of Parasitology, Kuvin Centre for the Study of Infectious and Tropical Diseases, The Hebrew University Hadassah- Medical School, Jerusalem, Israel. KEY WORDS: Lepidoptera, Israel, Levant, Nolidae, Eariadinae, Chloephorinae, phenology, ecology, host- plants. ÊËÞ×ÅÂÛÅ ÑËÎÂÀ: Lepidoptera, Èçðàèëü, Ëåâàíò, Nolidae, Eariadinae, Chloephorinae, ôåíîëîãèÿ, ýêîëîãèÿ, êîðìîâûå ðàñòåíèÿ. ABSTRACT: The distribution, flight period and âèä, Microxestis wutzdorffi (Püngeler, 1907), ñîáðàííûé abundance of six Israeli Eariadinae and eight Chloe- 80 ëåò íàçàä, íå îáíàðóæåí çà âðåìÿ ðàáîòû phorinae species (Noctuoidea, Nolidae) are summa- Èçðàèëüñêî-Ãåðìàíñêîãî Ïðîåêòà ïî èçó÷åíèþ Lepi- rized. Seven species are new records for Israel: Earias doptera. Äëÿ âñåõ âèäîâ ïðèâîäÿòñÿ äàííûå ïî biplaga Walker, 1866, Earias cupreoviridis (Walker, ÷èñëåííîñòè, ðàñïðåäåëåíèþ, ôåíîëîãèè è ýêîëîãèè. 1862), Acryophora dentula (Lederer, 1870), Bryophilop- Äëÿ ïÿòè âèäîâ âïåðâûå óêàçàíû êîðìîâûå ðàñòåíèÿ. sis roederi (Standfuss, 1892), Nycteola revayana (Sco- poli, 1772), Nycteola columbana (Turner, 1925) and Nycteola asiatica (Krulikovsky, 1904). Three species, Introduction E. biplaga E. cupreoviridis and N. revayana, are re- corded for the first time from the Levante. Only one The Nolidae is a family that has changed in its species, Microxestis wutzdorffi (Püngeler, 1907), col- coverage several times during the past. -

Biodiversity of Insect Pest Complex Infesting Okra [Abelmoschus
Journal of Entomology and Zoology Studies 2017; 5(5): 1968-1972 E-ISSN: 2320-7078 P-ISSN: 2349-6800 Biodiversity of insect pest complex infesting okra JEZS 2017; 5(5): 1968-1972 © 2017 JEZS [Abelmoschus esculentus] in Tripura, N.E. India Received: 11-07-2017 Accepted: 12-08-2017 Navendu Nair Navendu Nair, U Giri, T Bhattacharjee, B Thangjam, N Paul and MR Department of Agriculture Debnath Entomology, College of Agriculture, Tripura, India Abstract U Giri Field experiments were conducted to study the biodiversity of insect pest species infesting okra in Department of Agronomy, Tripura during two seasons viz, summer (April to July, 2016) and winter (October, 2016 to January, College of Agriculture, Tripura, 2017). A total of twenty eight insect pest species belonging to 6 orders were found to infest this crop. India Maximum number of species were belonging to Hemiptera (12 nos.) followed by Lepidoptera (9 nos.) and Coleoptera (4 nos.). The Shannon and Wiener diversity index (H’) during summer and winter season T Bhattacharjee for the insect pest complex of okra was calculated as 1.01 and 0.91, respectively. The Simpson’s Department of Horticulture, diversity index (D) during summer and winter season was calculated as 0.14 and 0.19, respectively. College of Agriculture, Tripura, India These two indices for summer and winter season were more or less equal and exhibited a similar diversification. Similarly, Species Richness (7.31 and 7.49) and Species Evenness (0.71 and 0.64) during B Thangjam summer and winter season were more or less equal. However, by number the Leaf hopper (Amrasca Department of Agriculture biguttula biguttula ), Leaf beetle (Podagrica sp.), Blister beetle (Mylabris pustulata) and Leaf folder Entomology, College of (Syllepte derogata) were more abundant in the field during summer season where as, Leaf hopper Agriculture, Tripura, India (Amrasca biguttula biguttula), Aphid (Aphis gossypii), White fly (Bemisia tabaci) and Leaf folder (Syllepte derogata) were more abundant during winter season. -

Redalyc.SECONDARY METABOLITES of the ANNONACEAE, SOLANACEAE and MELIACEAE FAMILIES USED AS BIOLOGICAL CONTROL of INSECTS
Tropical and Subtropical Agroecosystems E-ISSN: 1870-0462 [email protected] Universidad Autónoma de Yucatán México Castillo-Sánchez, Luis Enrique; Jiménez-Osornio, Juan José; Delgado-Herrera, María América SECONDARY METABOLITES OF THE ANNONACEAE, SOLANACEAE AND MELIACEAE FAMILIES USED AS BIOLOGICAL CONTROL OF INSECTS Tropical and Subtropical Agroecosystems, vol. 12, núm. 3, septiembre-diciembre, 2010, pp. 445-462 Universidad Autónoma de Yucatán Mérida, Yucatán, México Available in: http://www.redalyc.org/articulo.oa?id=93915170004 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Tropical and Subtropical Agroecosystems, 12 (2010): 445 -462 REVIEW [REVISIÓN] SECONDARY METABOLITES OF THE ANNONACEAE, SOLANACEAE Tropical and AND MELIACEAE FAMILIES USED AS BIOLOGICAL CONTROL OF INSECTS Subtropical [METABOLITOS SECUNDARIOS DE LAS FAMILIAS ANNONACEAE, SOLANACEAE Y MELIACEAE USADAS COMO CONTROL BIOLÓGICO Agroecosystems DE INSECTOS] Luis Enrique Castillo-Sánchez1*, Juan José Jiménez-Osornio2 and María América Delgado-Herrera3. 1Technological Institute of Tizimin 3.5 km final highway Cupul airport to Tizimin. Tizimin, Yucatan, Mexico. Email: [email protected] 2Tropical Natural Resources Management and Conservation Department, Biological Sciences and Animal Husbandry Campus, Autonomous University of Yucatan. -

Efficacy of Bt Proteins and the Effect of Temperature on the Development of Spiny Bollworm in South Africa
Efficacy of Bt proteins and the effect of temperature on the development of spiny bollworm in South Africa D Fourie 20670591 Dissertation submitted in fulfilment of the requirements for the degree Magister Scientiae in Environmental Sciences at the Potchefstroom Campus of the North-West University Supervisor: Prof MJ du Plessis Co-supervisor: Prof J van den Berg May 2016 Acknowledgement I would like to thank the Lord above for giving me the strength to finish this project, the love and passion I have for nature and the opportunity to do something I feel passionate about. Thank you Lord for the ability to study, and all the opportunities You send my way. With Your grace, anything is possible. I would like to thank my supervisor prof. Hannalene du Plessis for all her support, inspiration and guidance during this project, but particularly for her assistance in the statistical analysis. I learned a lot and appreciate all the time and effort we spent to finish this project. I would also like to thank my co-supervisor prof. Johnnie van den Berg for the support to finish this project and his assistance and time during the writing of this dissertation, I appreciate it a lot. I would like to thank all my friends for their help, support and encouragement during the time of this project. I am truly grateful for your help during field work and your assistance in the laboratory. A special thanks to Phillip Mphuthi for all his help in collecting Earias spp. moths. I appreciate all your help during the late hours and your hospitality during my stay in Rustenburg. -

Biological Control of Certain Insect Pests Attacking Cotton Plants in Sohag Governorate
201 Middle East Journal of Agriculture Research, 3(2): 201-207, 2014 ISSN 2077-4605 Biological Control of Certain Insect Pests Attacking Cotton Plants in Sohag Governorate 1Salman, A.M.A., 2Karaman, G.A., 3El-Zoghbey, A.A. and 3Mazeed, A.R.A. 1Plant Protection Dept., Fac. of Agric., Sohag Univ. 2Plant Protection Dept., Fac. of Agric., Minia Univ. 3Biological Control Dept., Plant Protection Institute, ARC. ABSTRACT The present investigations were carried out to study the efficiency of releasing the egg parasitoid, Trichogramma evanescens (West.) only or with Chrysoperla carnea Stephens., Coccinella undecimpunctata L. and Coccinella septempunctata L. against the pink bollworm, Pectinophora gossypiella (Saund.), and the spiny bollworm, Earias insulana (Boisd.), Also the effect on cotton yield was determined. Data revealed that all biological control treatments decreased significantly the population of both pink and spiny bollworms, All treatments induced significant increase in cotton yield compared with the untreated check. Also, numbers of diapausing larvae were significantly reduced by all treatments in both years. and investigations the efficiency of releasing Ch. carnea, C. undecimpunctata and C. septempunctata against the cotton aphid, Aphis gossypii Glover in cotton fields during 2010 and 2011 seasons, cotton aphid in treated plots compared with the untreated check. Key words: Biological control, Pectinophora gossypiella, Earias insulana, Aphis gossypii, cotton. Introduction Cotton, Gossypium barbadence L. is one of the most important economical crops in Egypt and all over the world where it is employed in several industrial productions i.e. ginning, textile, Food oil, soap, furniture and many other industries, as well as a source of foreign coin when exported to another countries, (Al-Shannaf, 2010). -

Pdf (641.45 K)
Egypt. Acad. J. Biolog. Sci., 13(1):47-57 (2020) Egyptian Academic Journal of Biological Sciences A. Entomology ISSN 1687- 8809 http://eajbsa.journals.ekb.eg/ Field Application of Bio-Insecticides On Spiny Bollworm, Earias insulana (Bosid.) On Cotton by Using Recent Low Volume Ground Spraying Equipment Rehab, A.A. Dar; Dalia, E. Lotfy and Hemat, Z. Moustafa Plant Protection Res. instit. Agric. Res. Center, Dokki, Giza. Email: [email protected] ______________________________________________________________ ARTICLE INFO ABSTRACT Article History Field experiments were carried out of about 11 kirats planted with (Giza Received:3/1/2020 86) cotton variety during seasons 2018 and 2019 on 15th July and 1st, 15th Accepted:29/1/2020 August in a field located at Qaha Research Station, Plant Protection Research ---------------------- Institute, Qalyoupia governorate. The selected area was split into 10 plots and Keywords: the control plot. Entomopathogenic fungi and a commercial formulation of Cotton, Earias Bacillus thuringiensis (Berliner) were sprayed by using Hand-held Hydraulic insulana , sprayer (Matabi) (56 L./fed.) and Economy Micron ULVA (15 L./Fed.) on Bacillus cotton field highly infested with cotton Spiny bollworm Earias insulana thuringiensis , larvae. All tested Bio-Insecticides revealed a significant negative influence on Earias insulana larvae. The most effective on reduction percentage of bolls Metarhizium infestation is Metarhizium anisopliae, Beauveria bassiana either isolates or anisopliae , (W. P.) with Economy Micron ULVA (15 L./Fed.) followed by Bacillus Beauveria thuringiensis (W.P.) with Hand-held Hydraulic sprayer (Matabi) (56 L./Fed.). bassiana , Low It could be recommended to use these Bio-Insecticides with ULV spraying Volume ground equipment with not less than (15L/Fed.). -

Keywords Genetic Profile of Earias Insulana (Boisd.) (Lepidoptera
Egypt. J. Plant Prot. Res. Inst. (2021), 4 (2): 341–351 Egyptian Journal of Plant Protection Research Institute www.ejppri.eg.net Genetic profile characterization of Earias insulana (Lepidoptera:Noctuidae) in Egypt Hemat, Z. Moustafa; Mervat, A. Kandil and El-Bassouiny, H. M. Plant Protection Research Institute, Agricultural Research Center, Dokki, Giza, Egypt. ARTICLE INFO Abstract: Article History Genetic profile of Earias insulana (Boisd.) Received: 28/4/2021 (Lepidoptera:Noctuidae) larvae was identified by two genetic Accepted: 29/ 6 /2021 techniques (SCoT and ISSR) using PCR. Seven SCoT primers (SCoT1, SCoT2, SCoT4, SCoT7, SCoT11, SCoT12 and SCoT15) Keywords resulted in twelve different bands with molecular weight between 975 and 165bp, while, five ISSR primers (49A, 49B, HB-10, HB-11 Earias insulana, genetic and HB-12) resulted in fifteen different bands with molecular weight profile, SCoT, ISSR, between 1630 and 175 bp. The genome profile characterization using peroxidase and SCoT technique for E. insulana was studies for the first time in the polyphenyl-oxidase present work. E. insulana field strains were collected from five okra isozymes. fields in different Governorates ; Behera, Alexandria, Menofia, Qaluobya and Sharkeya during season 2019 compared to the reared laboratory strain to investigate genetic study. Results of isozymes showed differences among larvae in five Governorates , polyphenyl oxidase was different in all tested larvae. Whereas, two bands were appeared in larvae from Alexandria governorate of peroxidase, while, larvae from Qaluobya and Sharkeya Governorates have only light different band as control (Laboratory). Introduction marker system called Start Codon Targeted The spiny bollworm (SBW) Earias (SCoT) Polymorphism (Collard and Mackill, insulana (Boisd.) (Lepidoptera: Noctuidae) is 2002) was developed based on the short one of the most dangerous pests attacking conserved region.