Genome-Wide Association Meta-Analysis Supports Genes Involved in Valve And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human Nonsense-Mediated RNA Decay Initiates Widely by Endonucleolysis and Targets Snorna Host Genes
Downloaded from genesdev.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes Søren Lykke-Andersen,1,4 Yun Chen,2,4 Britt R. Ardal,1 Berit Lilje,2 Johannes Waage,2,3 Albin Sandelin,2 and Torben Heick Jensen1 1Centre for mRNP Biogenesis and Metabolism, Department of Molecular Biology and Genetics, Aarhus University, Aarhus DK-8000, Denmark; 2The Bioinformatics Centre, Department of Biology and Biotech Research and Innovation Centre, University of Copenhagen, Copenhagen DK-2200, Denmark Eukaryotic RNAs with premature termination codons (PTCs) are eliminated by nonsense-mediated decay (NMD). While human nonsense RNA degradation can be initiated either by an endonucleolytic cleavage event near the PTC or through decapping, the individual contribution of these activities on endogenous substrates has remained unresolved. Here we used concurrent transcriptome-wide identification of NMD substrates and their 59–39 decay intermediates to establish that SMG6-catalyzed endonucleolysis widely initiates the degradation of human nonsense RNAs, whereas decapping is used to a lesser extent. We also show that a large proportion of genes hosting snoRNAs in their introns produce considerable amounts of NMD-sensitive splice variants, indicating that these RNAs are merely by-products of a primary snoRNA production process. Additionally, transcripts from genes encoding multiple snoRNAs often yield alternative transcript isoforms that allow for differential expression of individual coencoded snoRNAs. Based on our findings, we hypothesize that snoRNA host genes need to be highly transcribed to accommodate high levels of snoRNA production and that the expression of individual snoRNAs and their cognate spliced RNA can be uncoupled via alternative splicing and NMD. -

A Genome-Wide Association Study of a Coronary Artery Disease Risk Variant
Journal of Human Genetics (2013) 58, 120–126 & 2013 The Japan Society of Human Genetics All rights reserved 1434-5161/13 www.nature.com/jhg ORIGINAL ARTICLE A genome-wide association study of a coronary artery diseaseriskvariant Ji-Young Lee1,16, Bok-Soo Lee2,16, Dong-Jik Shin3,16, Kyung Woo Park4,16, Young-Ah Shin1, Kwang Joong Kim1, Lyong Heo1, Ji Young Lee1, Yun Kyoung Kim1, Young Jin Kim1, Chang Bum Hong1, Sang-Hak Lee3, Dankyu Yoon5, Hyo Jung Ku2, Il-Young Oh4, Bong-Jo Kim1, Juyoung Lee1, Seon-Joo Park1, Jimin Kim1, Hye-kyung Kawk1, Jong-Eun Lee6, Hye-kyung Park1, Jae-Eun Lee1, Hye-young Nam1, Hyun-young Park7, Chol Shin8, Mitsuhiro Yokota9, Hiroyuki Asano10, Masahiro Nakatochi11, Tatsuaki Matsubara12, Hidetoshi Kitajima13, Ken Yamamoto13, Hyung-Lae Kim14, Bok-Ghee Han1, Myeong-Chan Cho15, Yangsoo Jang3,17, Hyo-Soo Kim4,17, Jeong Euy Park2,17 and Jong-Young Lee1,17 Although over 30 common genetic susceptibility loci have been identified to be independently associated with coronary artery disease (CAD) risk through genome-wide association studies (GWAS), genetic risk variants reported to date explain only a small fraction of heritability. To identify novel susceptibility variants for CAD and confirm those previously identified in European population, GWAS and a replication study were performed in the Koreans and Japanese. In the discovery stage, we genotyped 2123 cases and 3591 controls with 521 786 SNPs using the Affymetrix SNP Array 6.0 chips in Korean. In the replication, direct genotyping was performed using 3052 cases and 4976 controls from the KItaNagoya Genome study of Japan with 14 selected SNPs. -

Oncogenic Inhibition by a Deleted in Liver Cancer Gene Requires Cooperation Between Tensin Binding and Rho-Specific Gtpase-Activating Protein Activities
Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities Xiaolan Qian*, Guorong Li*, Holly K. Asmussen*, Laura Asnaghi*, William C. Vass*, Richard Braverman*, Kenneth M. Yamada†, Nicholas C. Popescu‡, Alex G. Papageorge*, and Douglas R. Lowy*§ *Laboratory of Cellular Oncology and ‡Laboratory of Experimental Carcinogenesis, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892; and †Laboratory of Cell and Developmental Biology, National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 20892 Communicated by Ira Pastan, National Institutes of Health, Bethesda, MD, April 2, 2007 (received for review March 1, 2007) The three deleted in liver cancer genes (DLC1–3) encode Rho- The prototypic member, designated DLC1, is localized to GTPase-activating proteins (RhoGAPs) whose expression is fre- chromosome 8p21–22 in a region that is commonly deleted in quently down-regulated or silenced in a variety of human malig- hepatocellular carcinoma (5). Its expression is frequently down- nancies. The RhoGAP activity is required for full DLC-dependent regulated or silenced in various solid tumors and hematologic tumor suppressor activity. Here we report that DLC1 and DLC3 bind malignancies, predominantly by promoter methylation (6–13). to human tensin1 and its chicken homolog. The binding has been Ectopic reexpression in DLC1-deficient cancer cell lines can mapped to the tensin Src homology 2 (SH2) and phosphotyrosine suppress cell proliferation, induce apoptosis, and reduce tumor- binding (PTB) domains at the C terminus of tensin proteins. Distinct igenicity. The RhoGAP activity appears to be required for these DLC1 sequences are required for SH2 and PTB binding. -

Transcriptomic and Proteomic Profiling Provides Insight Into
BASIC RESEARCH www.jasn.org Transcriptomic and Proteomic Profiling Provides Insight into Mesangial Cell Function in IgA Nephropathy † † ‡ Peidi Liu,* Emelie Lassén,* Viji Nair, Celine C. Berthier, Miyuki Suguro, Carina Sihlbom,§ † | † Matthias Kretzler, Christer Betsholtz, ¶ Börje Haraldsson,* Wenjun Ju, Kerstin Ebefors,* and Jenny Nyström* *Department of Physiology, Institute of Neuroscience and Physiology, §Proteomics Core Facility at University of Gothenburg, University of Gothenburg, Gothenburg, Sweden; †Division of Nephrology, Department of Internal Medicine and Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan; ‡Division of Molecular Medicine, Aichi Cancer Center Research Institute, Nagoya, Japan; |Department of Immunology, Genetics and Pathology, Uppsala University, Uppsala, Sweden; and ¶Integrated Cardio Metabolic Centre, Karolinska Institutet Novum, Huddinge, Sweden ABSTRACT IgA nephropathy (IgAN), the most common GN worldwide, is characterized by circulating galactose-deficient IgA (gd-IgA) that forms immune complexes. The immune complexes are deposited in the glomerular mesangium, leading to inflammation and loss of renal function, but the complete pathophysiology of the disease is not understood. Using an integrated global transcriptomic and proteomic profiling approach, we investigated the role of the mesangium in the onset and progression of IgAN. Global gene expression was investigated by microarray analysis of the glomerular compartment of renal biopsy specimens from patients with IgAN (n=19) and controls (n=22). Using curated glomerular cell type–specific genes from the published literature, we found differential expression of a much higher percentage of mesangial cell–positive standard genes than podocyte-positive standard genes in IgAN. Principal coordinate analysis of expression data revealed clear separation of patient and control samples on the basis of mesangial but not podocyte cell–positive standard genes. -

Genetic Alterations of Protein Tyrosine Phosphatases in Human Cancers
Oncogene (2015) 34, 3885–3894 © 2015 Macmillan Publishers Limited All rights reserved 0950-9232/15 www.nature.com/onc REVIEW Genetic alterations of protein tyrosine phosphatases in human cancers S Zhao1,2,3, D Sedwick3,4 and Z Wang2,3 Protein tyrosine phosphatases (PTPs) are enzymes that remove phosphate from tyrosine residues in proteins. Recent whole-exome sequencing of human cancer genomes reveals that many PTPs are frequently mutated in a variety of cancers. Among these mutated PTPs, PTP receptor T (PTPRT) appears to be the most frequently mutated PTP in human cancers. Beside PTPN11, which functions as an oncogene in leukemia, genetic and functional studies indicate that most of mutant PTPs are tumor suppressor genes. Identification of the substrates and corresponding kinases of the mutant PTPs may provide novel therapeutic targets for cancers harboring these mutant PTPs. Oncogene (2015) 34, 3885–3894; doi:10.1038/onc.2014.326; published online 29 September 2014 INTRODUCTION tyrosine/threonine-specific phosphatases. (4) Class IV PTPs include Protein tyrosine phosphorylation has a critical role in virtually all four Drosophila Eya homologs (Eya1, Eya2, Eya3 and Eya4), which human cellular processes that are involved in oncogenesis.1 can dephosphorylate both tyrosine and serine residues. Protein tyrosine phosphorylation is coordinately regulated by protein tyrosine kinases (PTKs) and protein tyrosine phosphatases 1 THE THREE-DIMENSIONAL STRUCTURE AND CATALYTIC (PTPs). Although PTKs add phosphate to tyrosine residues in MECHANISM OF PTPS proteins, PTPs remove it. Many PTKs are well-documented oncogenes.1 Recent cancer genomic studies provided compelling The three-dimensional structures of the catalytic domains of evidence that many PTPs function as tumor suppressor genes, classical PTPs (RPTPs and non-RPTPs) are extremely well because a majority of PTP mutations that have been identified in conserved.5 Even the catalytic domain structures of the dual- human cancers are loss-of-function mutations. -

TERRA: Telomeric Repeat-Containing RNA
The EMBO Journal (2009) 28, 2503–2510 | & 2009 European Molecular Biology Organization | Some Rights Reserved 0261-4189/09 www.embojournal.org TTHEH E EEMBOMBO JJOURNALOURN AL Focus Review TERRA: telomeric repeat-containing RNA Brian Luke1,2 and Joachim Lingner1,2,* lytic processing of chromosome ends and the end replication problem. This shortening can be counteracted by the cellular 1EPFL-Ecole Polytechnique Fe´de´rale de Lausanne, ISREC-Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland and reverse-transcriptase telomerase, which uses an internal RNA 2‘Frontiers in Genetics’ National Center for Competence in Research moiety as a template for the synthesis of telomere repeats (NCCR), Geneva, Switzerland (Cech, 2004; Blackburn et al, 2006). Telomerase is regulated at individual chromosome ends through telomere-binding Telomeres, the physical ends of eukaryotic chromosomes, proteins to mediate telomere length homoeostasis; however, consist of tandem arrays of short DNA repeats and a large in humans, telomerase is expressed in most tissues only set of specialized proteins. A recent analysis has identified during the first weeks of embryogenesis (Ulaner and telomeric repeat-containing RNA (TERRA), a large non- Giudice, 1997). Repression of telomerase in somatic cells is coding RNA in animals and fungi, which forms an integral thought to result in a powerful tumour-suppressive function. component of telomeric heterochromatin. TERRA tran- Short telomeres that accumulate following an excessive scription occurs at most or all chromosome ends and it number of cell division cycles induce cellular senescence, is regulated by RNA surveillance factors and in response to and this counteracts the growth of pre-malignant lesions. -

In This Table Protein Name, Uniprot Code, Gene Name P-Value
Supplementary Table S1: In this table protein name, uniprot code, gene name p-value and Fold change (FC) for each comparison are shown, for 299 of the 301 significantly regulated proteins found in both comparisons (p-value<0.01, fold change (FC) >+/-0.37) ALS versus control and FTLD-U versus control. Two uncharacterized proteins have been excluded from this list Protein name Uniprot Gene name p value FC FTLD-U p value FC ALS FTLD-U ALS Cytochrome b-c1 complex P14927 UQCRB 1.534E-03 -1.591E+00 6.005E-04 -1.639E+00 subunit 7 NADH dehydrogenase O95182 NDUFA7 4.127E-04 -9.471E-01 3.467E-05 -1.643E+00 [ubiquinone] 1 alpha subcomplex subunit 7 NADH dehydrogenase O43678 NDUFA2 3.230E-04 -9.145E-01 2.113E-04 -1.450E+00 [ubiquinone] 1 alpha subcomplex subunit 2 NADH dehydrogenase O43920 NDUFS5 1.769E-04 -8.829E-01 3.235E-05 -1.007E+00 [ubiquinone] iron-sulfur protein 5 ARF GTPase-activating A0A0C4DGN6 GIT1 1.306E-03 -8.810E-01 1.115E-03 -7.228E-01 protein GIT1 Methylglutaconyl-CoA Q13825 AUH 6.097E-04 -7.666E-01 5.619E-06 -1.178E+00 hydratase, mitochondrial ADP/ATP translocase 1 P12235 SLC25A4 6.068E-03 -6.095E-01 3.595E-04 -1.011E+00 MIC J3QTA6 CHCHD6 1.090E-04 -5.913E-01 2.124E-03 -5.948E-01 MIC J3QTA6 CHCHD6 1.090E-04 -5.913E-01 2.124E-03 -5.948E-01 Protein kinase C and casein Q9BY11 PACSIN1 3.837E-03 -5.863E-01 3.680E-06 -1.824E+00 kinase substrate in neurons protein 1 Tubulin polymerization- O94811 TPPP 6.466E-03 -5.755E-01 6.943E-06 -1.169E+00 promoting protein MIC C9JRZ6 CHCHD3 2.912E-02 -6.187E-01 2.195E-03 -9.781E-01 Mitochondrial 2- -
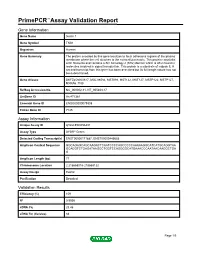
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name tensin 1 Gene Symbol TNS1 Organism Human Gene Summary The protein encoded by this gene localizes to focal adhesions regions of the plasma membrane where the cell attaches to the extracellular matrix. This protein crosslinks actin filaments and contains a Src homology 2 (SH2) domain which is often found in molecules involved in signal transduction. This protein is a substrate of calpain II. A second transcript from this gene has been described but its full length nature has not been determined. Gene Aliases DKFZp586K0617, MGC88584, MST091, MST122, MST127, MSTP122, MSTP127, MXRA6, TNS RefSeq Accession No. NC_000002.11, NT_005403.17 UniGene ID Hs.471381 Ensembl Gene ID ENSG00000079308 Entrez Gene ID 7145 Assay Information Unique Assay ID qHsaCED0056481 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000171887, ENST00000446688 Amplicon Context Sequence GGCAGAGCAGCAAGACTCAATCCCCAGCCCCCAAAAAGGCATCATGCAGGTAA GCAGGTCTGAGATAAGGCTGGTCCAGGGGCATGAAACCCAATAACAAGGCTGA G Amplicon Length (bp) 77 Chromosome Location 2:218668016-218668122 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 109 R2 0.9936 cDNA Cq 23.48 cDNA Tm (Celsius) 83 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 25.01 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report TNS1, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis -

Common Genetic Variants and Subclinical Atherosclerosis: the Multi-Ethnic Study of Atherosclerosis
Common Genetic Variants and Subclinical Atherosclerosis: The Multi-Ethnic Study of Atherosclerosis (MESA). Authors Authors: Jose D. Vargas, Ani Manichaikul, Xin Q. Wang, Stephen S. Rich, Jerome I. Rotter, Wendy S. Post, Joseph F. Polak, Matthew J. Budoff, and David A. Bluemke. Abstract Background: Subclinical atherosclerosis (sCVD), measured by coronary artery calcium (CAC) and carotid intima media thickness (CIMT) has been associated with cardiovascular disease (CVD). Genome Wide Association Studies (GWAS) of CVD have focused on Caucasian populations. We hypothesized that these associations would differ in populations from distinct genetic backgrounds. Methods and Results: The associations between sCVD and 66 single nucleotide polymorphisms (SNPs) from published GWAS of sCVD and CVD were tested in 8224 Multi-Ethnic Study of Atherosclerosis (MESA) and MESA Family participants (2685 Caucasians (EUA), 777 Chinese (CHN), 2588 African Americans (AFA), and 2174 Hispanic (HIS)) using an additive model adjusting for CVD risk factors, with SNP significance defined by a Bonferroni-corrected p < 7.6 x 10-4 (0.05/66). Results: In EUA there were significant associations with CAC in 9p21 (rs1333049, P=2 x 10-9; rs4977574, P= 4 x 10-9), COL4A1 (rs9515203, P=9 x 10-6), and PHACTR1 (rs9349379, P= 4 x 10-4). In HIS, SNPs were associated with CAC in 9p21 (rs1333049, P=8 x 10-5; rs4977574, P=5 x 10-5), APOA5 (rs964184, P=2 x 10-4), and ADAMTS7 (rs7173743, P=4 x 10-4). There were no associations with the 9p21 region in AFA and CHN. Fine mapping of the 9p21 region revealed SNPs with robust associations with CAC in EUA and HIS but no significant associations in AFA and CHN. -

Nº Ref Uniprot Proteína Péptidos Identificados Por MS/MS 1 P01024
Document downloaded from http://www.elsevier.es, day 26/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Nº Ref Uniprot Proteína Péptidos identificados 1 P01024 CO3_HUMAN Complement C3 OS=Homo sapiens GN=C3 PE=1 SV=2 por 162MS/MS 2 P02751 FINC_HUMAN Fibronectin OS=Homo sapiens GN=FN1 PE=1 SV=4 131 3 P01023 A2MG_HUMAN Alpha-2-macroglobulin OS=Homo sapiens GN=A2M PE=1 SV=3 128 4 P0C0L4 CO4A_HUMAN Complement C4-A OS=Homo sapiens GN=C4A PE=1 SV=1 95 5 P04275 VWF_HUMAN von Willebrand factor OS=Homo sapiens GN=VWF PE=1 SV=4 81 6 P02675 FIBB_HUMAN Fibrinogen beta chain OS=Homo sapiens GN=FGB PE=1 SV=2 78 7 P01031 CO5_HUMAN Complement C5 OS=Homo sapiens GN=C5 PE=1 SV=4 66 8 P02768 ALBU_HUMAN Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 66 9 P00450 CERU_HUMAN Ceruloplasmin OS=Homo sapiens GN=CP PE=1 SV=1 64 10 P02671 FIBA_HUMAN Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 58 11 P08603 CFAH_HUMAN Complement factor H OS=Homo sapiens GN=CFH PE=1 SV=4 56 12 P02787 TRFE_HUMAN Serotransferrin OS=Homo sapiens GN=TF PE=1 SV=3 54 13 P00747 PLMN_HUMAN Plasminogen OS=Homo sapiens GN=PLG PE=1 SV=2 48 14 P02679 FIBG_HUMAN Fibrinogen gamma chain OS=Homo sapiens GN=FGG PE=1 SV=3 47 15 P01871 IGHM_HUMAN Ig mu chain C region OS=Homo sapiens GN=IGHM PE=1 SV=3 41 16 P04003 C4BPA_HUMAN C4b-binding protein alpha chain OS=Homo sapiens GN=C4BPA PE=1 SV=2 37 17 Q9Y6R7 FCGBP_HUMAN IgGFc-binding protein OS=Homo sapiens GN=FCGBP PE=1 SV=3 30 18 O43866 CD5L_HUMAN CD5 antigen-like OS=Homo -

Discovery and Systematic Characterization of Risk Variants and Genes For
medRxiv preprint doi: https://doi.org/10.1101/2021.05.24.21257377; this version posted June 2, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY 4.0 International license . 1 Discovery and systematic characterization of risk variants and genes for 2 coronary artery disease in over a million participants 3 4 Krishna G Aragam1,2,3,4*, Tao Jiang5*, Anuj Goel6,7*, Stavroula Kanoni8*, Brooke N Wolford9*, 5 Elle M Weeks4, Minxian Wang3,4, George Hindy10, Wei Zhou4,11,12,9, Christopher Grace6,7, 6 Carolina Roselli3, Nicholas A Marston13, Frederick K Kamanu13, Ida Surakka14, Loreto Muñoz 7 Venegas15,16, Paul Sherliker17, Satoshi Koyama18, Kazuyoshi Ishigaki19, Bjørn O Åsvold20,21,22, 8 Michael R Brown23, Ben Brumpton20,21, Paul S de Vries23, Olga Giannakopoulou8, Panagiota 9 Giardoglou24, Daniel F Gudbjartsson25,26, Ulrich Güldener27, Syed M. Ijlal Haider15, Anna 10 Helgadottir25, Maysson Ibrahim28, Adnan Kastrati27,29, Thorsten Kessler27,29, Ling Li27, Lijiang 11 Ma30,31, Thomas Meitinger32,33,29, Sören Mucha15, Matthias Munz15, Federico Murgia28, Jonas B 12 Nielsen34,20, Markus M Nöthen35, Shichao Pang27, Tobias Reinberger15, Gudmar Thorleifsson25, 13 Moritz von Scheidt27,29, Jacob K Ulirsch4,11,36, EPIC-CVD Consortium, Biobank Japan, David O 14 Arnar25,37,38, Deepak S Atri39,3, Noël P Burtt4, Maria C Costanzo4, Jason Flannick40, Rajat M 15 Gupta39,3,4, Kaoru Ito18, Dong-Keun Jang4, -

Circrna Circ 102049 Implicates in Pancreatic Ductal Adenocarcinoma Progression Through Activating CD80 by Targeting Mir-455-3P
Hindawi Mediators of Inflammation Volume 2021, Article ID 8819990, 30 pages https://doi.org/10.1155/2021/8819990 Research Article circRNA circ_102049 Implicates in Pancreatic Ductal Adenocarcinoma Progression through Activating CD80 by Targeting miR-455-3p Jie Zhu,1 Yong Zhou,1 Shanshan Zhu,1 Fei Li,1 Jiajia Xu,1 Liming Zhang,1 and Hairong Shu 2 1Medical Laboratory, Taizhou Central Hospital (Taizhou University Hospital), Taizhou, Zhejiang, China 2Department of Medical Service, Taizhou Central Hospital (Taizhou University Hospital), Taizhou, Zhejiang, China Correspondence should be addressed to Hairong Shu; [email protected] Received 30 September 2020; Revised 27 November 2020; Accepted 13 December 2020; Published 7 January 2021 Academic Editor: Xiaolu Jin Copyright © 2021 Jie Zhu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Emerging evidence has shown that circular RNAs (circRNAs) and DNA methylation play important roles in the causation and progression of cancers. However, the roles of circRNAs and abnormal methylation genes in the tumorigenesis of pancreatic ductal adenocarcinoma (PDAC) are still largely unknown. Expression profiles of circRNA, gene methylation, and mRNA were downloaded from the GEO database, and differentially expressed genes were obtained via GEO2R, and a ceRNA network was constructed based on circRNA-miRNA pairs and miRNA-mRNA pairs. Inflammation-associated genes were collected from the GeneCards database. Then, functional enrichment analysis and protein-protein interaction (PPI) networks of inflammation- associated methylated expressed genes were investigated using Metascape and STRING databases, respectively, and visualized in Cytoscape.