UREA CYCLE Metabolic Pathway Dr
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Development of a System of High Ornithine and Citrulline Production by a Plant-Derived Lactic Acid Bacterium, Weissella Confusa K-28
Development of a system of high ornithine and citrulline production by a plant-derived lactic acid bacterium, Weissella confusa K-28 㸦᳜≀⏤᮶ங㓟⳦ :HLVVHOODFRQIXVD. ࠾ࡅࡿ ࢜ࣝࢽࢳࣥཬࡧࢩࢺࣝࣜࣥ㧗⏕⏘ࢩࢫࢸ࣒ࡢᵓ⠏㸧 Md Rakhimuzzaman Student ID: D164180 Supervisor: Prof. Masanori Sugiyama Department of Probiotic Science for Preventive Medicine Graduate School of Biomedical and Health Sciences Hiroshima University, Japan 2019 ABBREVIATIONS ADI arginine deiminase AUC area under curve BLAST basic local alignment search tool CK carbamate kinase dNTP deoxyribonucleotide triphosphate DDBJ DNA Data Bank of Japan EDTA ethylenediamine tetra-acetic acid EtOH ethanol EPS exopolysaccharide GABA gamma-aminobutyric acid GRAS generally recognized as safe HPLC high performance liquid chromatography LAB lactic acid bacteria Lb. Lactobacillus OD optical density OTC ornithine carbamoyltransferase PITC phenyl isothiocyanate TAE tris-acetate-ethylenediamine tetra-acetic acid TE tris-ethylenediamine tetra-acetic acid TEA triethylamine Tris tris(hydroxymethyl)aminomethane rDNA ribosomal deoxyribonucleic acid RT-PCR reverse transcription polymerase chain reaction CONTENTS INTRODUCTION 1-17 OBJECTIVES 18 MATERIALS AND METHODS 19-34 RESULTS 35-57 DISCUSSIONS 58-65 CONCLUSION 66-67 ACKNOWLEDGEMENTS 68 REFERENCES 69-84 INTRODUCTION Lactic acid bacteria (LAB) are an order of gram-positive, acid-tolerant, generally nonsporulating, non-respiring, either rod-shaped or spherical microorganisms. Since LAB lack the ability of synthesizing porphyrins (heme, and components of respiratory chains) (Pessione et al, 2010), the bacteria can not generate ATP without external heme supplementation (Pessione et al, 2010). The LAB strains can only obtain ATP by fermentation, usually they generate the ATP from some sugars. Because LAB do not use oxygen for energy production, the bacteria easily grow under anaerobic conditions, but they can also grow in presence of oxygen. -

Metabolic Reprogramming in Prostate Cancer
www.nature.com/bjc REVIEW ARTICLE Metabolic reprogramming in prostate cancer Fahim Ahmad 1,2, Murali Krishna Cherukuri2 and Peter L. Choyke1 Although low risk localised prostate cancer has an excellent prognosis owing to effective treatments, such as surgery, radiation, cryosurgery and hormone therapy, metastatic prostate cancer remains incurable. Existing therapeutic regimens prolong life; however, they are beset by problems of resistance, resulting in poor outcomes. Treatment resistance arises primarily from tumour heterogeneity, altered genetic signatures and metabolic reprogramming, all of which enable the tumour to serially adapt to drugs during the course of treatment. In this review, we focus on alterations in the metabolism of prostate cancer, including genetic signatures and molecular pathways associated with metabolic reprogramming. Advances in our understanding of prostate cancer metabolism might help to explain many of the adaptive responses that are induced by therapy, which might, in turn, lead to the attainment of more durable therapeutic responses. British Journal of Cancer https://doi.org/10.1038/s41416-021-01435-5 BACKGROUND Several tools have been designed to assess metabolism without Cellular metabolism involves complex biochemical processes disturbing the system. Advances in nuclear magnetic resonance through which specific nutrients are consumed. Carbohydrates, and mass spectroscopy have helped to quantify the carbon influx fatty acids and amino acids are the main source of nutrients for of the central metabolic pathways (e.g. TCA cycle, glycolysis and energy homeostasis and macromolecular synthesis. They are the pentose phosphate pathway) in mammalian systems, and this main constituent of core metabolic pathways that can be approach has been instrumental in demonstrating that the classified as anabolic, catabolic or waste producing (Fig. -

Urea Adducts of the Esters of Stearic Acid
University of the Pacific Scholarly Commons University of the Pacific Theses and Dissertations Graduate School 1952 Urea adducts of the esters of stearic acid Paul Elliott Greene University of the Pacific Follow this and additional works at: https://scholarlycommons.pacific.edu/uop_etds Part of the Chemistry Commons Recommended Citation Greene, Paul Elliott. (1952). Urea adducts of the esters of stearic acid. University of the Pacific, Thesis. https://scholarlycommons.pacific.edu/uop_etds/1186 This Thesis is brought to you for free and open access by the Graduate School at Scholarly Commons. It has been accepted for inclusion in University of the Pacific Theses and Dissertations by an authorized administrator of Scholarly Commons. For more information, please contact [email protected]. I': UREA,, ADDUCTS OF THE :ESTERS OF S'rEARIC ACID A Thesis Presented to Tb.~ Faculty of the Department of Chemistry College of the Pacific In Partial Fulfillment of the Requi:r>ements for the Degree Master of A:rts by Paul Elliott Greene... June 1952 ACKNOt~L1i:DGE~4ENT Appreciation is expressed to th.e faculty members of the Chemistry Department of the College of the Paciflc and ospecially to Dr. H;merson Cobb for the gu.idance and criticism given to the writer during this p~>r:i.od of' study. TABLE OF CONTENTS PAGE Urea and urea complexes .. • .. • • .. • .. • • • • • • 1 Adducts of' hydrocl.'ll1bon der1 vati ves • • • • • * • Similar molecular complexes • • • • • • • • • Structure cf ut.. ea £'.d.ducts • • . .. .. • 7 --------App-lioat:Lon-of_:ux~ee.-a.dd"u.cts-~--~-·--~-~-·-·-.. -·-·-· 11.---- P:r•eparati.on of intermedi.ates • • • • • • • • • • • 14 Purif:toation of ste:Htr:to ao:td . -

Amino Acid Degradation; Urea Cycle
Amino acid degradation; urea cycle Kristina Mlinac Jerković [email protected] In animals, amino acids undergo oxidative degradation in three different metabolic circumstances: 1. During the normal synthesis and degradation of cellular proteins, some amino acids that are released from protein breakdown and are not needed for new protein synthesis undergo oxidative degradation. 2. When a diet is rich in protein and the ingested amino acids exceed the body’s needs for protein synthesis, the surplus is catabolized; amino acids cannot be stored. 3. During starvation or in uncontrolled diabetes mellitus, when carbohydrates are either unavailable or not properly utilized, cellular proteins are used as fuel. Metabolic fates of amino groups - The metabolism of nitrogen-containing molecules such as proteins and nucleic acids differs significantly from that of carbohydrates and lipids. - Whereas the latter molecules can be stored and mobilized as needed for biosynthetic reactions or for energy generation, there is no nitrogen-storing molecule (one exception to this rule is storage protein in seeds). - Organisms must constantly replenish their supply of usable nitrogen to replace organic nitrogen that is lost in catabolism. For example, animals must have a steady supply of amino acids in their diets to replace the nitrogen excreted as urea, uric acid, and other nitrogenous waste products. - Since excess dietary amino acids are neither stored for future use nor excreted, they are converted to common metabolic intermediates such as pyruvate, oxaloacetate, acetyl-CoA, and α-keto-glutarate. - Consequently, amino acids are also precursors of glucose, fatty acids, and ketone bodies and are therefore metabolic fuels. -

Protein Metabolism and It's Disorders
Protein Metabolism and it’s Disorders (Course : M.Sc. Zoology) Course Code: ZOOL 4008 (Biochemistry and Metabolism), Semester –II Dr. Shyam Babu Prasad Assistant Professor Department of Zoology Mahatma Gandhi Central University (MGCU), Motihari-845401 (Bihar) Email: [email protected] Important facts of the Protein Metabolism •Proteins metabolism referred as synthesis of protein from Amino acids (Anabolism) and breakdown of proteins (Catabolism). •In the Unit II, we discussed the synthesis (Anabolism) of various form of the proteins from different amino acids. • Here we mainly focused on the Catabolism of the protein, which is integral part of metabolism of the nitrogen-containing molecules. • Nitrogen enters in to the body in various form of compounds that is present in food, the most important is amino acids (as dietary food). • During Protein/amino acid metabolism (Catabolism), Nitrogen leaves the body as urea, ammonia, and in form of other derivatives. • The Catabolism of the protein depends on the amino acid pool and protein turnover (the rate of protein synthesis and degradation is equal). • The amino acids present in through out of the body (cells, blood, and the extracellular fluids etc) is called amino acid pools. It is maintained by following factors as represented in below diagram: Body protein Dietary degradation Protein 400gm/day 600gm/day Supplied Amino acid pool Depleted Synthesis of Remaining Synthesis of Glucose, Synthesis of amino acids Nitrogen containing Glycogen, the Body burned as compound like Ketone Proteins -
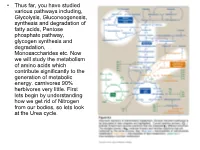
Transaminations and Urea Cycle
• Thus far, you have studied various pathways including, Glycolysis, Gluconeogenesis, synthesis and degradation of fatty acids, Pentose phosphate pathway, glycogen synthesis and degradation, Monosaccharides etc. Now we will study the metabolism of amino acids which contribute significantly to the generation of metabolic energy. carnivores 90% herbivores very little. First lets begin by understanding how we get rid of Nitrogen from our bodies, so lets look at the Urea cycle. 1 α Amino group keeps amino acids safe from oxidative breakdown. Removal of this group is essential for energy production it is an obligatory step for catabolism. Nitrogen can then be incorporated or excreted.This is done through transamination or oxidative deamination, provide amonia and aspartate sources for the Urea cycle. Transamination is funneled through glutamate. This is done through the transfer of the α amino group to α ketoglutarate. Product α-keto acid and glutamate. Alanine to pyruvate and aspartate to oxaloacetate all amino acids except lysine and threonina (lose the α amino through deamination. Glutamate dehydrogenase the oxidative deamination Glutamate only amino acid that undergoes rapid oxidative deamination (glutamate dehydrogenase NH3 + NADH), where amino groups are released as ammonia. After a meal rich in protein the levels of glutamate in liver are increased favoring amino acid degradation and formation of ammonia. Activators of this enzyme are ADP and GDP while ATP and GTP are inhibitors. This reaction occurs in the mitochondria. Ammonia can be transported to the liver from other tissues through two mechanisms that reduce the toxicity of ammonia. One through Glutamine (most tissues through, glutamine synthetase). In the liver the glutamine is converted back to glutamate (Fig. -

ATP Structure and Function
ATP: Universal Currency of Cellular Energy All living things including plants, animals, birds, insects, humans need energy for the proper functioning of cells, tissues and other organ systems. As we are aware that green plants, obtain their energy from the sunlight, and animals get their energy by feeding on these plants. Energy acts as a source of fuel. We, humans, gain energy from the food we eat, but how are the energy produced and stored in our body. A living cell cannot store significant amounts of free energy. Excess free energy would result in an increase of heat in the cell, which would result in excessive thermal motion that could damage and then destroy the cell. Rather, a cell must be able to handle that energy in a way that enables the cell to store energy safely and release it for use only as needed. Living cells accomplish this by using the compound adenosine triphosphate (ATP). ATP is often called the “energy currency” of the cell, and, like currency, this versatile compound can be used to fill any energy need of the cell. How? It functions similarly to a rechargeable battery. When ATP is broken down, usually by the removal of its terminal phosphate group, energy is released. The energy is used to do work by the cell, usually by the released phosphate binding to another molecule, activating it. For example, in the mechanical work of muscle contraction, ATP supplies the energy to move the contractile muscle proteins. Recall the active transport work of the sodium-potassium pump in cell membranes. -

UREA/AMMONIA (Rapid)
www.megazyme.com UREA/AMMONIA (Rapid) ASSAY PROCEDURE K-URAMR 04/20 (For the rapid assay of urea and ammonia in all samples, including grape juice and wine) (*50 Assays of each per Kit) * The number of tests per kit can be doubled if all volumes are halved © Megazyme 2020 INTRODUCTION: Urea and ammonia are widely occurring natural compounds. As urea is the most abundant organic solute in urine, and ammonia is produced as a consequence of microbial protein catabolism, these analytes serve as reliable quality indicators for food products such as fruit juice, milk, cheese, meat and seafood. Ammonium carbonate is used as a leaven in baked goods such as quick breads, cookies and muffins. Unlike some other kits, this kit benefits from the use of a glutamate dehydrogenase that is not inhibited by tannins found in, for example, grape juice and wine. In the wine industry, ammonia determination is important in the calculation of yeast available nitrogen (YAN). YAN is comprised of three highly variable components, free ammonium ions, primary amino nitrogen (from free amino acids) and the contribution from the sidechain of L-arginine.1 For the most accurate determination of YAN, all three components should be quantified, and this is possible using Megazyme’s L-Arginine/Urea/Ammonia Kit (K-LARGE) and NOPA Kit (K-PANOPA). Urea determination can be important in preventing the formation of the known carcinogen ethyl carbamate (EC) in finished wine. PRINCIPLE: Urea is hydrolysed to ammonia (NH3) and carbon dioxide (CO2) by the enzyme urease (1). (urease) (1) Urea + H2O 2NH3 + CO2 In the presence of glutamate dehydrogenase (GlDH) and reduced nicotinamide-adenine dinucleotide phosphate (NADPH), ammonia (as + ammonium ions; NH4 ) reacts with 2-oxoglutarate to form L-glutamic acid and NADP+ (2). -

Laboratory Diagnostic Approaches in Metabolic Disorders
470 Review Article on Inborn Errors of Metabolism Page 1 of 14 Laboratory diagnostic approaches in metabolic disorders Ruben Bonilla Guerrero1, Denise Salazar2, Pranoot Tanpaiboon2,3 1Formerly Quest Diagnostics, Inc., Ruben Bonilla Guerrero, Rancho Santa Margarita, CA, USA; 2Quest Diagnostics, Inc., Denise Salazar and Pranoot Tanpaiboon, San Juan Capistrano, CA, USA; 3Genetics and Metabolism, Children’s National Rare Disease Institute, Washington, DC, USA Contributions: (I) Conception and design: All authors; (II) Administrative support: R Bonilla Guerrero; (III) Provision of study materials or patients: All authors; (IV) Collection and assembly of data: All authors; (V) Data analysis and interpretation: None; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Ruben Bonilla Guerrero. Formerly Quest Diagnostics, Inc., Ruben Bonilla Guerrero, 508 Sable, Rancho Santa Margarita, CA 92688, USA. Email: [email protected]. Abstract: The diagnosis of inborn errors of metabolism (IEM) takes many forms. Due to the implementation and advances in newborn screening (NBS), the diagnosis of many IEM has become relatively easy utilizing laboratory biomarkers. For the majority of IEM, early diagnosis prevents the onset of severe clinical symptoms, thus reducing morbidity and mortality. However, due to molecular, biochemical, and clinical variability of IEM, not all disorders included in NBS programs will be detected and diagnosed by screening alone. This article provides a general overview and simplified guidelines for the diagnosis of IEM in patients with and without an acute metabolic decompensation, with early or late onset of clinical symptoms. The proper use of routine laboratory results in the initial patient assessment is also discussed, which can help guide efficient ordering of specialized laboratory tests to confirm a potential diagnosis and initiate treatment as soon as possible. -

Estonian Statistics on Medicines 2016 1/41
Estonian Statistics on Medicines 2016 ATC code ATC group / Active substance (rout of admin.) Quantity sold Unit DDD Unit DDD/1000/ day A ALIMENTARY TRACT AND METABOLISM 167,8985 A01 STOMATOLOGICAL PREPARATIONS 0,0738 A01A STOMATOLOGICAL PREPARATIONS 0,0738 A01AB Antiinfectives and antiseptics for local oral treatment 0,0738 A01AB09 Miconazole (O) 7088 g 0,2 g 0,0738 A01AB12 Hexetidine (O) 1951200 ml A01AB81 Neomycin+ Benzocaine (dental) 30200 pieces A01AB82 Demeclocycline+ Triamcinolone (dental) 680 g A01AC Corticosteroids for local oral treatment A01AC81 Dexamethasone+ Thymol (dental) 3094 ml A01AD Other agents for local oral treatment A01AD80 Lidocaine+ Cetylpyridinium chloride (gingival) 227150 g A01AD81 Lidocaine+ Cetrimide (O) 30900 g A01AD82 Choline salicylate (O) 864720 pieces A01AD83 Lidocaine+ Chamomille extract (O) 370080 g A01AD90 Lidocaine+ Paraformaldehyde (dental) 405 g A02 DRUGS FOR ACID RELATED DISORDERS 47,1312 A02A ANTACIDS 1,0133 Combinations and complexes of aluminium, calcium and A02AD 1,0133 magnesium compounds A02AD81 Aluminium hydroxide+ Magnesium hydroxide (O) 811120 pieces 10 pieces 0,1689 A02AD81 Aluminium hydroxide+ Magnesium hydroxide (O) 3101974 ml 50 ml 0,1292 A02AD83 Calcium carbonate+ Magnesium carbonate (O) 3434232 pieces 10 pieces 0,7152 DRUGS FOR PEPTIC ULCER AND GASTRO- A02B 46,1179 OESOPHAGEAL REFLUX DISEASE (GORD) A02BA H2-receptor antagonists 2,3855 A02BA02 Ranitidine (O) 340327,5 g 0,3 g 2,3624 A02BA02 Ranitidine (P) 3318,25 g 0,3 g 0,0230 A02BC Proton pump inhibitors 43,7324 A02BC01 Omeprazole -

Amino Acid Catabolism: Urea Cycle the Urea Bi-Cycle Two Issues
BI/CH 422/622 OUTLINE: OUTLINE: Protein Degradation (Catabolism) Digestion Amino-Acid Degradation Inside of cells Urea Cycle – dealing with the nitrogen Protein turnover Ubiquitin Feeding the Urea Cycle Activation-E1 Glucose-Alanine Cycle Conjugation-E2 Free Ammonia Ligation-E3 Proteosome Glutamine Amino-Acid Degradation Glutamate dehydrogenase Ammonia Overall energetics free Dealing with the carbon transamination-mechanism to know Seven Families Urea Cycle – dealing with the nitrogen 1. ADENQ 5 Steps 2. RPH Carbamoyl-phosphate synthetase oxidase Ornithine transcarbamylase one-carbon metabolism Arginino-succinate synthetase THF Arginino-succinase SAM Arginase 3. GSC Energetics PLP uses Urea Bi-cycle 4. MT – one carbon metabolism 5. FY – oxidases Amino Acid Catabolism: Urea Cycle The Urea Bi-Cycle Two issues: 1) What to do with the fumarate? 2) What are the sources of the free ammonia? a-ketoglutarate a-amino acid Aspartate transaminase transaminase a-keto acid Glutamate 1 Amino Acid Catabolism: Urea Cycle The Glucose-Alanine Cycle • Vigorously working muscles operate nearly anaerobically and rely on glycolysis for energy. a-Keto acids • Glycolysis yields pyruvate. – If not eliminated (converted to acetyl- CoA), lactic acid will build up. • If amino acids have become a fuel source, this lactate is converted back to pyruvate, then converted to alanine for transport into the liver. Excess Glutamate is Metabolized in the Mitochondria of Hepatocytes Amino Acid Catabolism: Urea Cycle Excess glutamine is processed in the intestines, kidneys, and liver. (deaminating) (N,Q,H,S,T,G,M,W) OAA à Asp Glutamine Synthetase This costs another ATP, bringing it closer to 5 (N,Q,H,S,T,G,M,W) 29 N 2 Amino Acid Catabolism: Urea Cycle Excess glutamine is processed in the intestines, kidneys, and liver. -

Catalytic Carbonylation of Amines And
CATALYTIC CARBONYLATION OF AMINES AND DIAMINES AS AN ALTERNATIVE TO PHOSGENE DERIVATIVES: APPLICATION TO SYNTHESES OF THE CORE STRUCTURE OF DMP 323 AND DMP 450 AND OTHER FUNCTIONALIZED UREAS By KEISHA-GAY HYLTON A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2004 Copyright 2004 by Keisha-Gay Hylton Dedicated to my father Alvest Hylton; he never lived to celebrate any of my achievements but he is never forgotten. ACKNOWLEDGMENTS A number of special individuals have contributed to my success. I thank my mother, for her never-ending support of my dreams; and my grandmother, for instilling integrity, and for her encouragement. Special thanks go to my husband Nemanja. He is my confidant, my best friend, and the love of my life. I thank him for providing a listening ear when I needed to “discuss” my reactions; and for his support throughout these 5 years. To my advisor (Dr. Lisa McElwee-White), I express my gratitude for all she has taught me over the last 4 years. She has shaped me into the chemist I am today, and has provided a positive role model for me. I am eternally grateful. I, of course, could never forget to mention my group members. I give special mention to Corey Anthony, for all the free coffee and toaster strudels; and for helping to keep the homesickness at bay. I thank Daniel for all the good gossip and lessons about France. I thank Yue Zhang for carbonylation discussions, and lessons about China.