Role of Cdx Factors in Early Mesodermal Fate Decisions Tanya E
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prospective Isolation of NKX2-1–Expressing Human Lung Progenitors Derived from Pluripotent Stem Cells
The Journal of Clinical Investigation RESEARCH ARTICLE Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells Finn Hawkins,1,2 Philipp Kramer,3 Anjali Jacob,1,2 Ian Driver,4 Dylan C. Thomas,1 Katherine B. McCauley,1,2 Nicholas Skvir,1 Ana M. Crane,3 Anita A. Kurmann,1,5 Anthony N. Hollenberg,5 Sinead Nguyen,1 Brandon G. Wong,6 Ahmad S. Khalil,6,7 Sarah X.L. Huang,3,8 Susan Guttentag,9 Jason R. Rock,4 John M. Shannon,10 Brian R. Davis,3 and Darrell N. Kotton1,2 2 1Center for Regenerative Medicine, and The Pulmonary Center and Department of Medicine, Boston University School of Medicine, Boston, Massachusetts, USA. 3Center for Stem Cell and Regenerative Medicine, Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center, Houston, Texas, USA. 4Department of Anatomy, UCSF, San Francisco, California, USA. 5Division of Endocrinology, Diabetes and Metabolism, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, Massachusetts, USA. 6Department of Biomedical Engineering and Biological Design Center, Boston University, Boston, Massachusetts, USA. 7Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts, USA. 8Columbia Center for Translational Immunology & Columbia Center for Human Development, Columbia University Medical Center, New York, New York, USA. 9Department of Pediatrics, Monroe Carell Jr. Children’s Hospital, Vanderbilt University, Nashville, Tennessee, USA. 10Division of Pulmonary Biology, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA. It has been postulated that during human fetal development, all cells of the lung epithelium derive from embryonic, endodermal, NK2 homeobox 1–expressing (NKX2-1+) precursor cells. -

Reciprocal Regulation of TEAD4 and CCN2 for the Trophectoderm Development of the Bovine Blastocyst
155 6 REPRODUCTIONRESEARCH Reciprocal regulation of TEAD4 and CCN2 for the trophectoderm development of the bovine blastocyst Hiroki Akizawa1,2, Ken Kobayashi3, Hanako Bai1, Masashi Takahashi1, Shinjiro Kagawa1, Hiroaki Nagatomo1,† and Manabu Kawahara1 1Laboratory of Animal Genetics and Reproduction, Research Faculty of Agriculture, Hokkaido University, Sapporo, Japan, 2Japan Society for the Promotion of Science (JSPS Research Fellow), Tokyo, Japan and 3Laboratory of Cell and Tissue Biology, Research Faculty of Agriculture, Hokkaido University, Sapporo, Japan Correspondence should be addressed to M Kawahara; Email: [email protected] †(Hiroaki Nagatomo is now at Department of Biotechnology, Faculty of Life and Environmental Science, University of Yamanashi, Kofu, Japan) Abstract The first segregation at the blastocyst stage is the symmetry-breaking event to characterize two cell components; namely, inner cell mass (ICM) and trophectoderm (TE). TEA domain transcription factor 4 (TEAD4) is a well-known regulator to determine TE properties of blastomeres in rodent models. However, the roles of bovine TEAD4 in blastocyst development have been unclear. We here aimed to clarify the mechanisms underlining TE characterization by TEAD4 in bovine blastocysts. We first found that the TEAD4 mRNA expression level was greater in TE than in ICM, which was further supported by TEAD4 immunofluorescent staining. Subsequently, we examined the expression patterns of TE-expressed genes; CDX2, GATA2 and CCN2, in the TEAD4-knockdown (KD) blastocysts. These expression levels significantly decreased in the TEAD4 KD blastocysts compared with controls. Of these downregulated genes, the CCN2 expression level decreased the most. We further analyzed the expression levels of TE-expressed genes; CDX2, GATA2 and TEAD4 in the CCN2 KD blastocysts. -

Regulation of the Tumor Suppressor Homeogene Cdx2 by Hnf4a in Intestinal Cancer
Oncogene (2013) 32, 3782–3788 & 2013 Macmillan Publishers Limited All rights reserved 0950-9232/13 www.nature.com/onc SHORT COMMUNICATION Regulation of the tumor suppressor homeogene Cdx2 by HNF4a in intestinal cancer T Saandi1,2, F Baraille3,4,5, L Derbal-Wolfrom1,2, A-L Cattin3,4,5, F Benahmed1,6, E Martin1,2, P Cardot3,4,5, B Duclos1,2,7, A Ribeiro3,4,5, J-N Freund1,2,8 and I Duluc1,2,8 The gut-specific homeotic transcription factor Cdx2 is a crucial regulator of intestinal development and homeostasis, which is downregulated in colorectal cancers (CRC) and exhibits a tumor suppressor function in the colon. We have previously established that several endodermal transcription factors, including HNF4a and GATA6, are involved in Cdx2 regulation in the normal gut. Here we have studied the role of HNF4a in the mechanism of deregulation of Cdx2 in colon cancers. Crossing ApcD14/ þ mice prone to spontaneous intestinal tumor development with pCdx2-9LacZ transgenic mice containing the LacZ reporter under the control of the 9.3-kb Cdx2 promoter showed that this promoter segment contains sequences recapitulating the decrease of Cdx2 expression in intestinal cancers. Immunohistochemistry revealed that HNF4a, unlike GATA6, exhibited a similar decrease to Cdx2 in genetic (Apcmin/ þ and ApcD14/ þ ) and chemically induced (Azoxymethane (AOM) treatment) models of intestinal tumors in mice. HNF4a and Cdx2 also exhibited a comparable deregulated pattern in human CRC. Correlated patterns were observed between HNF4a and Cdx2 in several experimental models of human colon cancer cell lines: xenografts in nude mice, wound healing and glucose starvation. -

CDX2 As a Useful Marker of Colorectal Adenocarcinoma Metastases to Lung in Pre-Operative Biopsy Specimens
87-92 1/6/07 12:56 Page 87 ONCOLOGY REPORTS 18: 87-92, 2007 87 CDX2 as a useful marker of colorectal adenocarcinoma metastases to lung in pre-operative biopsy specimens SHIGEBUMI TANAKA1, KANA SAITO1,2, TOMOKAZU ITO1, KOHEI TAJIMA3, AKIRA MOGI3, YOSHINORI SHITARA4, TAKAAKI SANO2 and HIROYUKI KUWANO1 Departments of 1General Surgical Science, 2Tumor Pathology, Gunma Graduate School of Medicine, 3-39-22 Showa-machi, Maebashi, Gunma 371-8511; 3Department of Surgery, Fujioka General Hospital, 942-1 Fujioka, Fujioka, Gunma 375-8503; 4Department of Surgery, Isesaki General Hospital, 12-1 Tsunatorihoncho, Isesaki, Gunma 372-0821, Japan Received January 12, 2007; Accepted April 17, 2007 Abstract. Although distinguishing metastatic colorectal as a solitary nodule. Differential diagnosis between metastatic adenocarcinoma from primary lung adenocarcinoma is often adenocarcinoma of colorectal origin and primary lung adeno- difficult, pre- or intra-operative identification is very important, carcinoma using imaging techniques such as chest computed as the resection areas for each diagnosis differ substantially. tomography (CT) is thus often difficult in patients with a CDX2, a recently cloned homeobox gene, represents a highly history of colorectal cancer. However, this differentiation is specific and sensitive marker of colorectal adenocarcinoma. crucial for therapeutic and prognostic purposes (1). Particularly We evaluated CDX2 expression using pre- and intra-operative in cases with operative indications, pre- or intra-operative biopsy specimens. The study examined 50 consecutive colo- identification is very important, as the resection areas differ rectal adenocarcinoma metastases to the lung, including 20 substantially between the diagnoses. If the tumor is a solitary biopsy specimens and 66 resected specimens, and 21 primary metastatic colorectal carcinoma to the lung, partial lung lung adenocarcinomas. -

Expression Pattern of the Class I Homeobox Genes in Ovarian Carcinoma
J Gynecol Oncol Vol. 21, No. 1:29-37, March 2010 DOI:10.3802/jgo.2010.21.1.29 Original Article Expression pattern of the class I homeobox genes in ovarian carcinoma Jin Hwa Hong1, Jae Kwan Lee1, Joong Jean Park2, Nak Woo Lee1, Kyu Wan Lee1, Jung Yeol Na1 Departments of 1Obstetrics and Gynecology, 2Physiology, Korea University College of Medicine, Seoul, Korea Objective: Although some sporadic reports reveal the link between the homeobox (HOX) genes and ovarian carcinoma, there is no comprehensive analysis of the expression pattern of the class I homeobox genes in ovarian carcinoma that determines the candidate genes involved in ovarian carcinogenesis. Methods: The different patterns of expression of 36 HOX genes were analyzed, including 4 ovarian cancer cell lines and 4 normal ovarian tissues. Using a reverse transcription-polymerase chain reaction (RT-PCR) and quantification analysis, the specific gene that showed a significantly higher expression in ovarian cancer cell lines than in normal ovaries was selected, and western blot analysis was performed adding 7 ovarian cancer tissue specimens. Finally, immunohistochemical and immunocytochemical analyses were performed to compare the pattern of expression of the specific HOX gene between ovarian cancer tissue and normal ovaries. Results: Among 36 genes, 11 genes had a different level of mRNA expression between the cancer cell lines and the normal ovarian tissues. Of the 11 genes, only HOXB4 had a significantly higher level of expression in ovarian cancer cell lines than in normal ovaries (p=0.029). Based on western blot, immunohistochemical, and immunocytochemical analyses, HOXB4 was expressed exclusively in the ovarian cancer cell lines or cancer tissue specimens, but not in the normal ovaries. -

Nutritional Rickets: Vitamin D, Calcium, and the Genetic Make-Up
nature publishing group Articles Translational Investigation Nutritional rickets: vitamin D, calcium, and the genetic make-up Mohamed El Kholy1, Heba Elsedfy1, Monica Fernández-Cancio2, Rasha Tarif Hamza1, Nermine Hussein Amr1, Alaa Youssef Ahmed1, Nadin Nabil Toaima1 and Laura Audí2 BACKGROUND: The prevalence of vitamin D (vitD) deficiency pigmentation are therefore main determinants of circulat- presenting as rickets is increasing worldwide. Insufficient sun ing vitD levels (2) that can be increased through dietary vitD exposure, vitD administration, and/or calcium intake are the intake (3). Interindividual differences in aspects of the vitD main causes. However, vitD system-related genes may also endocrine system have been well documented. They could be have a role. caused by genetic differences in important proteins in the vitD METHODS: Prospective study: 109 rachitic children com- endocrine system, such as VDR (4–6). pleted a 6-mo study period or until rachitic manifestations Multiple polymorphic variations exist in the VDR gene: disappeared. Thirty children were selected as controls. Clinical 5’-promoter variations can affect mRNA expression pat- and biochemical data were evaluated at baseline in patients terns and levels while 3’-UTR sequence variations can affect and controls and biochemistry re-evaluated at radiological mRNA stability and/or protein translation efficiency (7–10). healing. Therapy was stratified in three different protocols. Other genes encoding proteins in the vitD endocrine sys- Fifty-four single-nucleotide polymorphisms (SNPs) of five vitD tem (25-hydroxylase (CP2R1), 1-α-hydroxylase (CYP27B1), system genes (VDR, CP2R1, CYP27B1, CYP24A1, and GC) were 24-hydroxylase (CYP24A1), and vitamin D binding globulin genotyped and their association with clinical and biochemcial (GC)) also present polymorphic variations that could influ- data was analyzed. -

Changes in Gene Expression During the Early to Mid-Luteal (Receptive Phase) Transition in Human Endometrium Detected by High-Density Microarray Screening
Molecular Human Reproduction Vol.8, No.9 pp. 871–879, 2002 Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening Daniel D.Carson1,4, Errin Lagow1, Amantha Thathiah1, Rania Al-Shami1, Mary C.Farach-Carson1, Michael Vernon2, Lingwen Yuan3, Marc A.Fritz3 and Bruce Lessey3 1Department of Biological Sciences, University of Delaware, Newark, DE 19716, 2UNC Neuroscience Center and 3Department of Obstetrics and Gynecology, University of North Carolina, Chapel Hill, NC 27599–0001, USA 4To whom correspondence should be addressed. E-mail: [email protected] High density cDNA microarray screening was used to determine changes in gene expression occurring during the transition between the early luteal (prereceptive) and mid-luteal (receptive) phases in human endometrium. Of ~12 000 genes profiled, 693 (5.8%) displayed >2-fold differences in relative levels of expression between these stages. Of these, 370 genes (3.1%) displayed decreases ranging from 2- to >100-fold while 323 genes (2.7%) displayed increases ranging from 2- to >45-fold. Many genes correspond to mRNAs encoding proteins previously shown to change in a similar manner between the proliferative and mid-luteal phases, serving as one validation of the microarray screening results. In addition, novel genes were identified. Genes encoding cell surface receptors, adhesion and extracellular matrix proteins and growth factors accounted for 20% of the changes. Several genes were studied further by Northern blot analyses. These results confirmed that claudin-4/Clostridium perfringens enterotoxin (CPE) receptor and osteopontin (OPN) mRNA increased ~4- and 12-fold respectively, while betaig-H3 (BIGH3) decreased >80% during the early to mid-luteal transition. -

Deregulation of the HOXA10 Homeobox Gene in Endometrial Carcinoma: Role in Epithelial-Mesenchymal Transition
Research Article Deregulation of the HOXA10 Homeobox Gene in Endometrial Carcinoma: Role in Epithelial-Mesenchymal Transition Hiroyuki Yoshida,1 Russell Broaddus,2 Wenjun Cheng,1 SuSu Xie,2 and Honami Naora1 Departments of 1Molecular Therapeutics and 2Pathology, The University of Texas M.D. Anderson Cancer Center, Houston, Texas Abstract normally expressed during early urogenital development, and is expressed in renal tumors (6). PAX2 is thought to promote cell Homeobox genes encode transcription factors that control cell survival, and inhibiting PAX2 expression in cancer cells induces differentiation and play essential roles in developmental apoptosis (7). On the other hand, many homeobox genes that are patterning. Increasing evidence indicates that many homeo- normally expressed in differentiated tissues are lost or down- box genes are aberrantly expressed in cancers, and that their regulated in tumors. Nkx3.1 controls prostate morphogenesis and deregulation significantly contributes to tumor progression. is normally expressed in the prostate during development and in The homeobox gene HOXA10 controls uterine organogenesis the adult (8). NKX3.1 is frequently deleted in prostate cancers (9), during embryonic development and functional endometrial and enforced expression of NKX3.1 in prostate cancer cells differentiation in the adult. We investigated whether HOXA10 inhibits cell growth (10). Many other homeobox genes contribute expression is deregulated in endometrial carcinomas, and to tumor progression by promoting cell growth and/or survival how counteracting this aberrant expression modifies tumor (4, 5). However, given the important role of homeobox genes in behavior. We found that down-regulation of HOXA10 expres- cell differentiation, surprisingly little is known regarding their role sion in endometrial carcinomas strongly correlates with in EMT. -

Cdx2 Protein Expression in Normal and Malignant Human Tissues: an Immunohistochemical Survey Using Tissue Microarrays Christopher A
Cdx2 Protein Expression in Normal and Malignant Human Tissues: An Immunohistochemical Survey Using Tissue Microarrays Christopher A. Moskaluk, M.D., Ph.D., Hong Zhang, M.D., Steven M. Powell, M.D., Lisa A. Cerilli, M.D., Garret M. Hampton, Ph.D., Henry F. Frierson, Jr., M.D. Departments of Pathology (CAM, LAC, HFF), of Biochemistry and Molecular Genetics (CAM), and of Medicine (SMP), University of Virginia Health System, Charlottesville, Virginia; Department of Pathology (HZ), Anhui Medical University, Hefei, China; and Genomics Institute of the Novartis Research Foundation (GMH), San Diego, California KEY WORDS: Cdx2, immunohistochemistry, RNA Cdx2 has been identified as a marker of colon can- profiling, Tissue microarray. cer in RNA-profiling experiments. We show here Mod Pathol 2003;16(9):913–919 that the detection of Cdx2 protein by immunohis- tochemistry correlates well with RNA transcript lev- Cdx2 is a homeobox gene that has been shown to els as detected by oligonucleotide microarrays. Us- play a role in the development of the small and ing tissue microarrays containing most normal large intestine in mammals and in the differentia- tissue types and an antibody to the Cdx2 protein, tion of intestinal epithelial cells. The Cdx2 gene is strong diffuse Cdx2 staining was only seen in the expressed in all but the most distal portions of the nuclei of small and large intestinal epithelium and intestinal tract during development (1, 2). In a cell portions of the pancreatic duct system. In tissue culture model, Cdx2 has been shown to decrease microarrays containing 745 cancers from many an- cellular replication and induce differentiation to atomic sites, colonic adenocarcinomas showed mature intestinal epithelium (3). -

The Homeobox Gene CDX2 Is Aberrantly Expressed and Associated with an Inferior Prognosis in Patients with Acute Lymphoblastic Leukemia
Leukemia (2009) 23, 649–655 & 2009 Macmillan Publishers Limited All rights reserved 0887-6924/09 $32.00 www.nature.com/leu ORIGINAL ARTICLE The homeobox gene CDX2 is aberrantly expressed and associated with an inferior prognosis in patients with acute lymphoblastic leukemia S Thoene1,2,4, VPS Rawat1,2,4, B Heilmeier1, E Hoster1, KH Metzeler1, T Herold1, W Hiddemann1,2,NGo¨kbuget3, D Hoelzer3, SK Bohlander1,2, M Feuring-Buske1,2 and C Buske1,2 1Department of Medicine III, Klinikum Grosshadern, Munich, Germany; 2Clinical Cooperative Group Leukemia, Helmholtz Center Munich for Environmental Health, Munich, Germany and 3Department of Medicine III, Johann Wolfgang Goethe University, Frankfurt/Main, Germany Molecular characterization of acute lymphoblastic leukemia gene deregulation.11 However, it was shown that besides Hox (ALL) has greatly improved the ability to categorize and genes Cdx2 regulates other stem cell regulatory genes, such as Scl, prognostify patients with this disease. In this study, we show 12 that the proto-oncogene CDX2 is aberrantly expressed in the Gata1 and Runx1. Thus, aberrant expression of CDX2 might majority of cases with B-lineage ALL and T-ALL. High expres- perturb the stem cell regulatory network at different levels. sion of CDX2 correlated significantly with the ALL subtype We now report that CDX2 is aberrantly expressed in 81% of pro-B ALL, cALL, Ph þ ALL and early T-ALL. Furthermore, high adult patients with ALL, and that high expression levels of this expression of CDX2 was associated with inferior overall proto-oncogene predict poor treatment outcome in patients with survival and showed up as a novel and strong risk factor for this disease. -

Regulation of Trophoblast-Specific Factors by GATA2 and GATA3 In
Journal of Reproduction and Development, Vol. 57, No. 4, 2011, 10-186K —Original Article— Regulation of Trophoblast-Specific Factors by GATA2 and GATA3 in Bovine Trophoblast CT-1 Cells Hanako BAI1), Toshihiro SAKURAI1), Yohhei SOMEYA1), Toshihiro KONNO1), Atsushi IDETA2), Yoshito AOYAGI2) and Kazuhiko IMAKAWA1) 1)Laboratory of Animal Breeding, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657 and 2)Zen-noh ET Center, Hokkaido 080-1407, Japan Abstract. Numerous transcription factors that regulate trophoblast developmental processes have been identified; however, the regulation of trophoblast-specific gene expression has not been definitively characterized. While a new role of Gata3 in trophoblast development was being demonstrated in mice, we examined effects of GATA transcription factors on conceptus interferon tau (IFNT), a major trophectoderm factor in ruminants. In this study, expression patterns of trophoblast ASCL2, CDX2, CSH1, ELF5, HAND1, IFNT, and TKDP1 mRNAs were initially examined, from which ASCL2, CDX2, IFNT, and TKDP1 mRNAs were found to be similar to those of GATA2 and GATA3 in days 17, 20, and 22 (day 0=day of estrus) bovine conceptuses. A chromatin immunoprecipitation (ChIP) assay revealed that endogenous GATA2 and GATA3 occupied GATA binding sites on the upstream regions of CSH1, IFNT, and TKDP1 genes and on the intron 1 region of CDX2 gene in bovine trophoblast CT-1 cells. In transient transfection analyses of the upstream region of bovine CSH1, and IFNT or the intron 1 region of CDX2 gene, over-expression of GATA2 induced transactivation of these trophoblast-specific genes in bovine non-trophoblast ear fibroblast EF cells, but over-expression of GATA3 did not substantially affect their transactivation. -
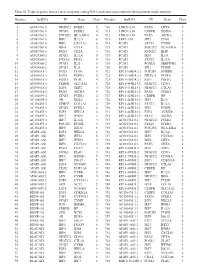
Table SI. Triple Negative Breast Cancer Long Non‑Coding RNA‑Mediated Transcriptional Dysregulation Triplet Network
Table SI. Triple negative breast cancer long non‑coding RNA‑mediated transcriptional dysregulation triplet network. Number lncRNA TF Gene Class Number lncRNA TF Gene Class 1 AC010761.8 NFATC2 FCER2 5 710 LINC01139 PAX5 CIITA 5 2 AC010761.8 STAT6 FCER2 4 711 LINC01139 CEBPB DDIT4 6 3 AC010761.8 POU2F2 HLA‑DRA 4 712 LINC01139 PAX5 AICDA 3 4 AC010761.8 IRF7 IL12A 6 713 LEF1‑AS1 SPI1 CD68 5 5 AC010761.8 IRF1 CXCL10 6 714 PCAT1 STAT1 TYMP 4 6 AC010761.8 IRF4 CIITA 5 715 PCAT1 POU2F2 HLA‑DRA 5 7 AC010761.8 PAX5 CIITA 4 716 PCAT1 RUNX2 IBSP 3 8 AC024560.2 STAT1 IL12A 5 717 PCAT1 IRF1 IL10 5 9 AC024560.2 FOXA2 PDX1 4 718 PCAT1 STAT2 IL12A 4 10 AC024560.2 STAT1 IL21 5 719 PCAT1 FOXD1 SERPINE1 3 11 AC024560.2 STAT2 IL21 5 720 PCAT1 SPI1 PTGIR 6 12 AC093818.1 PAX5 CD19 5 721 RP11‑169D4.2.1 POU2F1 GNRHR 3 13 AC093818.1 PAX5 FCER2 5 722 RP11‑169D4.2.1 NKX2‑5 PLOD1 4 14 AC093818.1 FOSL1 PLAU 4 723 RP11‑169D4.2.1 E2F1 POLE2 6 15 AC093818.1 CEBPB SLC11A1 6 724 RP11‑445K13.2 NFATC2 CD3G 5 16 AC093818.1 E2F1 TERT 3 725 RP11‑445K13.2 NFATC2 CTLA4 5 17 AC093818.1 PAX5 AICDA 5 726 RP11‑445K13.2 PAX5 FCER2 5 18 AC104699.1 STAT6 FCER2 4 727 RP11‑445K13.2 CEBPB HP 3 19 AC104699.1 AR KLK4 4 728 RP11‑445K13.2 IRF1 IL10 5 20 AC156455.1 CEBPZ COL1A1 2 729 RP11‑445K13.2 STAT1 IL12A 5 21 AC156455.1 STAT6 FCER2 4 730 RP11‑445K13.2 SPI1 PTGIR 6 22 AC156455.1 E2F1 HELLS 6 731 RP11‑445K13.2 E2F4 AURKB 6 23 AC156455.1 IRF7 IFNB1 3 732 RP11‑445K13.2 PAX5 AICDA 5 24 AC156455.1 IRF7 IL12A 3 733 AC010761.8.1 NFATC2 FCER2 5 25 AC156455.1 POU2F1 PTGIR 3 734 AC010761.8.1 STAT6