Fermentative Microbes of Khadi, a Traditional Alcoholic Beverage of Botswana
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Social Life of Khadi: Gandhi's Experiments with the Indian
The Social Life of Khadi: Gandhi’s Experiments with the Indian Economy, c. 1915-1965 by Leslie Hempson A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (History) in the University of Michigan 2018 Doctoral Committee: Associate Professor Farina Mir, Co-Chair Professor Mrinalini Sinha, Co-Chair Associate Professor William Glover Associate Professor Matthew Hull Leslie Hempson [email protected] ORCID iD: 0000-0001-5195-1605 © Leslie Hempson 2018 DEDICATION To my parents, whose love and support has accompanied me every step of the way ii TABLE OF CONTENTS DEDICATION ii LIST OF FIGURES iv LIST OF ACRONYMS v GLOSSARY OF KEY TERMS vi ABSTRACT vii INTRODUCTION 1 CHAPTER 1: THE AGRO-INDUSTRIAL DIVIDE 23 CHAPTER 2: ACCOUNTING FOR BUSINESS 53 CHAPTER 3: WRITING THE ECONOMY 89 CHAPTER 4: SPINNING EMPLOYMENT 130 CONCLUSION 179 APPENDIX: WEIGHTS AND MEASURES 183 BIBLIOGRAPHY 184 iii LIST OF FIGURES FIGURE 2.1 Advertisement for a list of businesses certified by AISA 59 3.1 A set of scales with coins used as weights 117 4.1 The ambar charkha in three-part form 146 4.2 Illustration from a KVIC album showing Mother India cradling the ambar 150 charkha 4.3 Illustration from a KVIC album showing giant hand cradling the ambar charkha 151 4.4 Illustration from a KVIC album showing the ambar charkha on a pedestal with 152 a modified version of the motto of the Indian republic on the front 4.5 Illustration from a KVIC album tracing the charkha to Mohenjo Daro 158 4.6 Illustration from a KVIC album tracing -

Suicide Deaths and Quality of Indian Cotton: Perspectives from History of Technology and Khadi Movement Author(S): C
Suicide Deaths and Quality of Indian Cotton: Perspectives from History of Technology and Khadi Movement Author(s): C. Shambu Prasad Source: Economic and Political Weekly, Vol. 34, No. 5 (Jan. 30 - Feb. 5, 1999), pp. PE12-PE21 Published by: Economic and Political Weekly Stable URL: http://www.jstor.org/stable/4407604 . Accessed: 13/06/2014 06:16 Your use of the JSTOR archive indicates your acceptance of the Terms & Conditions of Use, available at . http://www.jstor.org/page/info/about/policies/terms.jsp . JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. Economic and Political Weekly is collaborating with JSTOR to digitize, preserve and extend access to Economic and Political Weekly. http://www.jstor.org This content downloaded from 130.92.9.57 on Fri, 13 Jun 2014 06:16:17 AM All use subject to JSTOR Terms and Conditions Suicide Deaths and Quality of Indian Cotton Perspectives from History of Technology and Khadi Movement C Shambu Prasad The suicide deaths of farmers is a failure of agricultural science and the historical nature of the crisis needs to be appreciated. This paper seeks to retrace the route by which the present connections between Indian cotton and the mechanised textile industry were first established, a direction that has led to the present crisis on the fields of the cotton jflrmers. -

Evaluation Study on Khadi and Village Industries Programme
Evaluation Study on Khadi and Village Industries Programme Programme Evaluation Organisation Planning Commission Government of India New Delhi-110001 March 2001 1 CONTENTS Preface i Executive Summary iii I. CHAPTER 1. Background - Khadi and Village Industries Programme ………. 1-4 I. Introduction ……………………………………………………. 1 II. Special Employment Programme for 50 Selected Districts through KVI …….. 2 III. Implementation Methodology ……………………………………………….. 2 IV. Need for the Study ……………………………………………………. 3 2. Evaluation Study – Objectives and Methodology ……………..………… 5-9 2.2.1 Objective of the Evaluation Study…………………………………………… 5 2.3 Methodology ………………………………………………………………. 6 2.4.1 Instruments ………………………………………………………………. 6 2.5.1 Selection of Sample …………………………………………………………… 7 2.6.1 Selection of Beneficiaries ………………………………………………… 7 2.7 Selection of Individual Beneficiaries (Employees/Key Persons) …………….. 7 2.8 Coverage ……………………………………………………………… 8 2.9 Reference Period ……………………………………………………… 8 2.10 Orientation of the Field Teams ………………………………………….. 8 Annexure – I …………………………………………………………………………… 9 3. Administration - Planning, Implementation and Monitoring…….. 10-18 3.1 Administrative Structure …………………………………………………… 10 3.2 Directorate of REGP …… …………………………………………………… 10 3.3 KVI Boards …………………………………………………………………. 11 3.4 District Administration ……………………………………………………… 11 3.5 Coordination between KVIC & KVIB ……………………………………. 12 3.6 Planning ……………………………………………………………………. 12 3.7 Implementation ……………………………………………………………… 13 3.8 Certification Committee ……………………………………………………. 14 3.9 Quality -

The Big Governance Issues in Botswana
MARCH 2021 THE BIG GOVERNANCE ISSUES IN BOTSWANA A CIVIL SOCIETY SUBMISSION TO THE AFRICAN PEER REVIEW MECHANISM Contents Executive Summary 3 Acknowledgments 7 Acronyms and Abbreviations 8 What is the APRM? 10 The BAPS Process 12 Ibrahim Index of African Governance Botswana: 2020 IIAG Scores, Ranks & Trends 120 CHAPTER 1 15 Introduction CHAPTER 2 16 Human Rights CHAPTER 3 27 Separation of Powers CHAPTER 4 35 Public Service and Decentralisation CHAPTER 5 43 Citizen Participation and Economic Inclusion CHAPTER 6 51 Transparency and Accountability CHAPTER 7 61 Vulnerable Groups CHAPTER 8 70 Education CHAPTER 9 80 Sustainable Development and Natural Resource Management, Access to Land and Infrastructure CHAPTER 10 91 Food Security CHAPTER 11 98 Crime and Security CHAPTER 12 108 Foreign Policy CHAPTER 13 113 Research and Development THE BIG GOVERNANCE ISSUES IN BOTSWANA: A CIVIL SOCIETY SUBMISSION TO THE APRM 3 Executive Summary Botswana’s civil society APRM Working Group has identified 12 governance issues to be included in this submission: 1 Human Rights The implementation of domestic and international legislation has meant that basic human rights are well protected in Botswana. However, these rights are not enjoyed equally by all. Areas of concern include violence against women and children; discrimination against indigenous peoples; child labour; over reliance on and abuses by the mining sector; respect for diversity and culture; effectiveness of social protection programmes; and access to quality healthcare services. It is recommended that government develop a comprehensive national action plan on human rights that applies to both state and business. 2 Separation of Powers Political and personal interests have made separation between Botswana’s three arms of government difficult. -
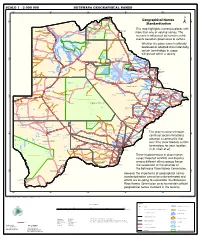
Geographical Names Standardization BOTSWANA GEOGRAPHICAL
SCALE 1 : 2 000 000 BOTSWANA GEOGRAPHICAL NAMES 20°0'0"E 22°0'0"E 24°0'0"E 26°0'0"E 28°0'0"E Kasane e ! ob Ch S Ngoma Bridge S " ! " 0 0 ' ' 0 0 ° Geographical Names ° ! 8 !( 8 1 ! 1 Parakarungu/ Kavimba ti Mbalakalungu ! ± n !( a Kakulwane Pan y K n Ga-Sekao/Kachikaubwe/Kachikabwe Standardization w e a L i/ n d d n o a y ba ! in m Shakawe Ngarange L ! zu ! !(Ghoha/Gcoha Gate we !(! Ng Samochema/Samochima Mpandamatenga/ This map highlights numerous places with Savute/Savuti Chobe National Park !(! Pandamatenga O Gudigwa te ! ! k Savu !( !( a ! v Nxamasere/Ncamasere a n a CHOBE DISTRICT more than one or varying names. The g Zweizwe Pan o an uiq !(! ag ! Sepupa/Sepopa Seronga M ! Savute Marsh Tsodilo !(! Gonutsuga/Gonitsuga scenario is influenced by human-centric Xau dum Nxauxau/Nxaunxau !(! ! Etsha 13 Jao! events based on governance or culture. achira Moan i e a h hw a k K g o n B Cakanaca/Xakanaka Mababe Ta ! u o N r o Moremi Wildlife Reserve Whether the place name is officially X a u ! G Gumare o d o l u OKAVANGO DELTA m m o e ! ti g Sankuyo o bestowed or adopted circumstantially, Qangwa g ! o !(! M Xaxaba/Cacaba B certain terminology in usage Nokaneng ! o r o Nxai National ! e Park n Shorobe a e k n will prevail within a society a Xaxa/Caecae/Xaixai m l e ! C u a n !( a d m a e a a b S c b K h i S " a " e a u T z 0 d ih n D 0 ' u ' m w NGAMILAND DISTRICT y ! Nxai Pan 0 m Tsokotshaa/Tsokatshaa 0 Gcwihabadu C T e Maun ° r ° h e ! 0 0 Ghwihaba/ ! a !( o 2 !( i ata Mmanxotae/Manxotae 2 g Botet N ! Gcwihaba e !( ! Nxharaga/Nxaraga !(! Maitengwe -

The Products That Buyers Are Actually Buying
WHAt’s SELLING NOW THE prODucts thAT buYErs ARE ActuAllY buYING Denim Denim Knitwear and Yarn n Cotton concept with n Textured, marble- Demin-look yarn in super softness and is effect denim from 71% cotton/29% rope dyed from Artistic Tat Fung Textiles Ltd. polyester from Denim (Pakistan) (Hong Kong) has an Winning Textiles Ltd. MOQ: 8,000 yds (flexible authentic vintage look (Hong Kong) Price: US$2.8-3.6/yd with a very soft hand. MOQ: in-stock This is a soft, heavy Price: US$7.2/kg “We use special spinnng 11.4 oz cotton denim from 71 cotton 29 polyester techniques and mix dif- 100 cotton 6oz denim weight (11.4 oz) Tat Fung (Hong Kong) “Cotton is very important. from Winning from Artistic Fabric Mills denim in 98% cot- It’s a natural fiber and ferent cotton, plus play (Pakistan) with the fabric finishing ton/2% elastane, that offers comfort stretch. buyers like it. Adding poly- to achieve the special hand feel “Brands like this fabric because ester gives the yarn more stabil- – this what buyers really like it is very soft and works for the ity. Armani Jeans and many US about it.” new silhouettes that are looser customers are buying this.” — Henry Wong fitting, like boyfriend jeans.” — Samson Lam, director at Artistic Denim — Sam Chan, sales manager Knitwear Denim Knitwear and Yarn n Hybrid Chino, which and Yarn n Glam shadow offers ‘denim with Chino n Mohair blends yarn from Huafu Top sennsibility’ in a two-ply from Best Leader Dyed Mélange Yarn construction. -

3.App-EFFECT of MERCERIZATION UNDER TENSION on THE
IMPACT: International Journal of Research in Applied, Natural and Social Sciences (IMPACT: IJRANSS) ISSN(P): 2347-4580; ISSN(E): 2321-8851 Vol. 5, Issue 4, Apr 2017, 19-26 © Impact Journals EFFECT OF MERCERIZATION UNDER TENSION ON THE DRAPABILITY AND STRENGTH OF COTTON KHADI FABRIC ADYA TIWARI 1 & RAJKUMARI JAIN 2 1Department of Textiles & Apparel Designing, College of Home Science, MPUAT, Udaipur, India 2Department of Clothing & Textiles, College of Home Science, Banasthali University, Rajasthan, India ABSTRACT Present study was aimed at assessing the effect of mercerization under tension on physical properties (drapability and tensile strength) of gray cotton khadi fabric. Different parameters of mercerization treatment such as conc. of sodium hydroxide palates and time duration of the treatment were optimized. Two methods of the treatment were given (slack mercerization and mercerization under tension) at 40 °C with different concentrations (5%, 15% and 25%) for different time periods (10min, 20min and 30min). The treated cotton khadi fabric was evaluated in terms of tensile strength and drapability of cotton khadi fabric. Mercerization under tension gave the best results with 25% concentration of NaOH for 30min at 40 °C followed by the fabric, with different time (10min and 20min) duration of the treatment. The results showed that as the time and concentrations were increased, the tensile strength was also increased of mercerization under tension as compare to the controlled and slack mercerization samples. Some properties were decreased after mercerization treatment for example %drape co-efficient was decreased and drape area also decreased means andanother mode of subjective expression of drapability i.e. higher the no. -

KHADI Emma Tarlo “This Is Sacred Cloth.” M. K. Gandhi Khadi Or
KHADI Emma Tarlo “This is sacred cloth.” M. K. Gandhi Khadi or khaddar is the term conventionally used in North and Central India to refer to varieties of coarse cotton cloth hand woven using hand spun yarn. This was the cloth commonly worn by peasant and artisan groups in pre-industrial India. It was made from locally grown cotton which would be harvested by peasants and labourers, spun by local women and woven into cloth by men from various specialist weaving castes. The precise technology involved in the production of khadi would vary from region to region, as would the techniques used for its decoration (dying, embroidery, printing etc.) Although hand spun hand woven cotton cloth of this kind was common throughout India, it was not until the early 20th century, when its production and use were in severe decline that the term “khadi” entered nationalist vocabulary and the cloth became a key visual symbol of India’s struggle from colonial rule. The effectiveness of khadi as a visual symbol of the Indian freedom struggle cannot be understood without examination of the critical role played by M. K. Gandhi (known by many as Mahatma – Great Soul) in elevating it to the status of a national cloth imbued with quasi-sacred properties. Gandhi’s success lay in his capacity to pick up, embody and develop existing political and economic critiques of colonialism and rework these through his own clothing practices and through his elaboration of the symbolism of cloth – a simple everyday material form to which people from all backgrounds could relate. -

Republic of Botswana the Project for Enhancing National Forest Monitoring System for the Promotion of Sustainable Natural Resource Management
DEPARTMENT OF FORESTRY AND RANGE RESOURCES (DFRR) MINISTRY OF ENVIRONMENT, NATURAL RESOURCES CONSERVATION AND TOURISM (MENT) REPUBLIC OF BOTSWANA REPUBLIC OF BOTSWANA THE PROJECT FOR ENHANCING NATIONAL FOREST MONITORING SYSTEM FOR THE PROMOTION OF SUSTAINABLE NATURAL RESOURCE MANAGEMENT PROJECT COMPLETION REPORT DECEMBER 2017 JAPAN INTERNATIONAL COOPERATION AGENCY(JICA) ORIENTAL CONSULTANTS GLOBAL CO., LTD. JAPAN FOREST TECHNOLOGY ASSOCIATION GE JR 17-131 DEPARTMENT OF FORESTRY AND RANGE RESOURCES (DFRR) MINISTRY OF ENVIRONMENT, NATURAL RESOURCES CONSERVATION AND TOURISM (MENT) REPUBLIC OF BOTSWANA REPUBLIC OF BOTSWANA THE PROJECT FOR ENHANCING NATIONAL FOREST MONITORING SYSTEM FOR THE PROMOTION OF SUSTAINABLE NATURAL RESOURCE MANAGEMENT PROJECT COMPLETION REPORT DECEMBER 2017 JAPAN INTERNATIONAL COOPERATION AGENCY(JICA) ORIENTAL CONSULTANTS GLOBAL CO., LTD. JAPAN FOREST TECHNOLOGY ASSOCIATION DFRR/JICA: Botswana Forest Distribution Map Zambia Angola Zambia Legend KASANE Angola ! ! Settlement CountryBoundary Riparian Forest Typical Forest Woodland Zimbabwe Zimbabwe Bushland/Shrubland Savanna/Grassland/Forbs MAUN ! NATA Baregorund ! TUTUME ! Desert/Sand Dunes Marsh/Wetland FRANCISTOWN Waterbody/Pan ! ORAPA Namibia ! TONOTA ! GHANZI Angola Zambia Namibia ! SELEBI-PHIKWE BOBONONG ! ! Zimbabwe SEROWE ! PALAPYE ! Namibia MAHALAPYE ! South Africa KANG ! MOLEPOLOLE MOCHUDI ! ! JWANENG ! GABORONE ! ´ 0 50 100 200 RAMOTSWA ! KANYE Kilometres ! Coordinate System: GCS WGS 1984 Datum: WGS 1984 LOBATSE ! Botswana Forest Distribution Map Produced from -

NEW AGE KHADI Khadi Is the Only Indian Feelgood Fabric As It Gives Employment to Thousands As Well As Boosts the Economy and Sustains Indigenous Artisans
Arup Datta NEW AGE KHADI Khadi is the only Indian feelgood fabric as it gives employment to thousands as well as boosts the economy and sustains indigenous artisans. Supporting khadi is one way of encouraging the talented artisan to live in his ancestral village rather than give up in despair and flock to an urban slum for an alternative employment. But khadi is far from fading away, thanks to the fillip given to the fabric by the Prime Minister himself. MEHER CASTELINO writes on the state of khadi affairs and how some designers are moulding it anew. he name may have changed over Gandhi chose khadi as a symbol of his dreams the centuries but the weft and for India when he returned from South Africa. the warp have not. Khadi, as we The charkha was selected by him as a sign of call the handwoven fabric made non-violence and self-sufficiency and the material legendary by Mahatma Gandhi, woven from it – khadi – epitomised the nation’s Thas been around since times immemorial. Its feelings of patriotism and nationalism. The revival timeline is its own hurrah: among the greatest of the charkha was symbolic of the nation’s quest achievements of the Harappa and Mohenjo-Daro for freedom and self-reliance. civilisations were the mastery over hand-spinning In 1921, Gandhi thought of a strategy and and hand-weaving. Every pre-Aryan home had came up with the charkha as an icon of the its own charkha or spinning wheel. Invaders struggle for India’s freedom. That revived the came and went but khadi wove its way through moribund hand-spinning and hand-weaving the Vedic period, the Mughal and Medieval ages. -

Table of Contents
1 Table of Contents 2 Letter Words .................................................................................................................................2 3 Letter Words .................................................................................................................................3 4 Letter Words .................................................................................................................................5 5 Letter Words ...............................................................................................................................12 6 Letter Words ...............................................................................................................................25 7 Letter Words ...............................................................................................................................43 8 Letter Words ...............................................................................................................................60 All words are taken from OWL 22 HOW TO USE THIS DOCUMENT Have you ever wanted to maximize your studying time? Just buzzing through word lists do not ensure that you will ever play the word….ever. The word lists in this document were run through 917,607 full game simulations. Only words that were played at least 100 times are in this list and in the order of most frequently played. These lists are in order or probability to play with the first word being the most probable. To maximize the use of this list is easy. Simply -
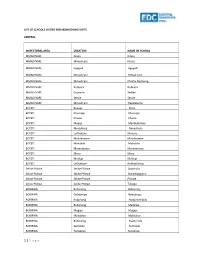
List of Schools Visited for Monitoring Visits
LIST OF SCHOOLS VISITED FOR MONITORING VISITS CENTRAL INSPECTORAL AREA LOCATION NAME OF SCHOOL MMADINARE Diloro Diloro MMADINARE Mmadinare Kelele MMADINARE Kgagodi Kgagodi MMADINARE Mmadinare Mmadinare MMADINARE Mmadinare Phethu Mphoeng MMADINARE Robelela Robelela MMADINARE Gojwane Sedibe MMADINARE Serule Serule MMADINARE Mmadinare Tlapalakoma BOTETI Rakops Etsile BOTETI Khumaga Khumaga BOTETI Khwee Khwee BOTETI Mopipi Manthabakwe BOTETI Mmadikola Mmadikola BOTETI Letlhakane Mokane BOTETI Mokoboxane Mokoboxane BOTETI Mokubilo Mokubilo BOTETI Moreomaoto Moreomaoto BOTETI Mosu Mosu BOTETI Motlopi Motlopi BOTETI Letlhakane Retlhatloleng Selibe Phikwe Selibe Phikwe Boitshoko Selibe Phikwe Selibe Phikwe Boswelakgomo Selibe Phikwe Selibe Phikwe Phikwe Selibe Phikwe Selibe Phikwe Tebogo BOBIRWA Bobonong Bobonong BOBIRWA Gobojango Gobojango BOBIRWA Bobonong Mabumahibidu BOBIRWA Bobonong Madikwe BOBIRWA Mogapi Mogapi BOBIRWA Molalatau Molalatau BOBIRWA Bobonong Rasetimela BOBIRWA Semolale Semolale BOBIRWA Tsetsebye Tsetsebye 1 | P a g e MAHALAPYE WEST Bonwapitse Bonwapitse MAHALAPYE WEST Mahalapye Leetile MAHALAPYE WEST Mokgenene Mokgenene MAHALAPYE WEST Moralane Moralane MAHALAPYE WEST Mosolotshane Mosolotshane MAHALAPYE WEST Otse Setlhamo MAHALAPYE WEST Mahalapye St James MAHALAPYE WEST Mahalapye Tshikinyega MHALAPYE EAST Mahalapye Flowertown MHALAPYE EAST Mahalapye Mahalapye MHALAPYE EAST Matlhako Matlhako MHALAPYE EAST Mmaphashalala Mmaphashalala MHALAPYE EAST Sefhare Mmutle PALAPYE NORTH Goo-Sekgweng Goo-Sekgweng PALAPYE NORTH Goo-Tau Goo-Tau