Endosymbiosis, a Proven Theory Or Evolution Myth?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pinpointing the Origin of Mitochondria Zhang Wang Hanchuan, Hubei
Pinpointing the origin of mitochondria Zhang Wang Hanchuan, Hubei, China B.S., Wuhan University, 2009 A Dissertation presented to the Graduate Faculty of the University of Virginia in Candidacy for the Degree of Doctor of Philosophy Department of Biology University of Virginia August, 2014 ii Abstract The explosive growth of genomic data presents both opportunities and challenges for the study of evolutionary biology, ecology and diversity. Genome-scale phylogenetic analysis (known as phylogenomics) has demonstrated its power in resolving the evolutionary tree of life and deciphering various fascinating questions regarding the origin and evolution of earth’s contemporary organisms. One of the most fundamental events in the earth’s history of life regards the origin of mitochondria. Overwhelming evidence supports the endosymbiotic theory that mitochondria originated once from a free-living α-proteobacterium that was engulfed by its host probably 2 billion years ago. However, its exact position in the tree of life remains highly debated. In particular, systematic errors including sparse taxonomic sampling, high evolutionary rate and sequence composition bias have long plagued the mitochondrial phylogenetics. This dissertation employs an integrated phylogenomic approach toward pinpointing the origin of mitochondria. By strategically sequencing 18 phylogenetically novel α-proteobacterial genomes, using a set of “well-behaved” phylogenetic markers with lower evolutionary rates and less composition bias, and applying more realistic phylogenetic models that better account for the systematic errors, the presented phylogenomic study for the first time placed the mitochondria unequivocally within the Rickettsiales order of α- proteobacteria, as a sister clade to the Rickettsiaceae and Anaplasmataceae families, all subtended by the Holosporaceae family. -

Origin and Evolution of Eukaryotic Compartmentalization
TESI DOCTORAL UPF 2016 Origin and evolution of eukaryotic compartmentalization Alexandros Pittis Director Dr. Toni Gabaldon´ Comparative Genomics Group Bioinformatics and Genomics Department Centre for Genomic Regulation (CRG) To my father Stavros for the PhD he never did Acknowledgments Looking back at the years of my PhD in the Comparative Genomics group at CRG, I feel it was equally a process of scientific, as well as personal development. All this time I was very privileged to be surrounded by people that offered me their generous support in both fronts. And they were available in the very moments that I was fighting more myself than the unsolvable (anyway) comparative genomics puzzles. I hope that in the future I will have many times the chance to express them my appreciation, way beyond these few words. First, to Toni, my supervisor, for all his support, and patience, and confidence, and respect, especially at the moments that things did not seem that promising. He offered me freedom, to try, to think, to fail, to learn, to achieve that few enjoy during their PhD, and I am very thankful to him. Then, to my good friends and colleagues, those that I found in the group already, and others that joined after me. A very special thanks to Marinita and Jaime, for all their valuable time, and guidance and paradigm, which nevertheless I never managed to follow. They have both marked my phylogenomics path so far and I cannot escape. To Les and Salvi, my fellow students and pals at the time for all that we shared; to Gab and Fran and Dam, for -

Virus World As an Evolutionary Network of Viruses and Capsidless Selfish Elements
Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Koonin, E. V., & Dolja, V. V. (2014). Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements. Microbiology and Molecular Biology Reviews, 78(2), 278-303. doi:10.1128/MMBR.00049-13 10.1128/MMBR.00049-13 American Society for Microbiology Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Eugene V. Koonin,a Valerian V. Doljab National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland, USAa; Department of Botany and Plant Pathology and Center for Genome Research and Biocomputing, Oregon State University, Corvallis, Oregon, USAb Downloaded from SUMMARY ..................................................................................................................................................278 INTRODUCTION ............................................................................................................................................278 PREVALENCE OF REPLICATION SYSTEM COMPONENTS COMPARED TO CAPSID PROTEINS AMONG VIRUS HALLMARK GENES.......................279 CLASSIFICATION OF VIRUSES BY REPLICATION-EXPRESSION STRATEGY: TYPICAL VIRUSES AND CAPSIDLESS FORMS ................................279 EVOLUTIONARY RELATIONSHIPS BETWEEN VIRUSES AND CAPSIDLESS VIRUS-LIKE GENETIC ELEMENTS ..............................................280 Capsidless Derivatives of Positive-Strand RNA Viruses....................................................................................................280 -

Evidence Supporting an Antimicrobial Origin of Targeting Peptides to Endosymbiotic Organelles
cells Article Evidence Supporting an Antimicrobial Origin of Targeting Peptides to Endosymbiotic Organelles Clotilde Garrido y, Oliver D. Caspari y , Yves Choquet , Francis-André Wollman and Ingrid Lafontaine * UMR7141, Institut de Biologie Physico-Chimique (CNRS/Sorbonne Université), 13 Rue Pierre et Marie Curie, 75005 Paris, France; [email protected] (C.G.); [email protected] (O.D.C.); [email protected] (Y.C.); [email protected] (F.-A.W.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 19 June 2020; Accepted: 24 July 2020; Published: 28 July 2020 Abstract: Mitochondria and chloroplasts emerged from primary endosymbiosis. Most proteins of the endosymbiont were subsequently expressed in the nucleo-cytosol of the host and organelle-targeted via the acquisition of N-terminal presequences, whose evolutionary origin remains enigmatic. Using a quantitative assessment of their physico-chemical properties, we show that organelle targeting peptides, which are distinct from signal peptides targeting other subcellular compartments, group with a subset of antimicrobial peptides. We demonstrate that extant antimicrobial peptides target a fluorescent reporter to either the mitochondria or the chloroplast in the green alga Chlamydomonas reinhardtii and, conversely, that extant targeting peptides still display antimicrobial activity. Thus, we provide strong computational and functional evidence for an evolutionary link between organelle-targeting and antimicrobial peptides. Our results support the view that resistance of bacterial progenitors of organelles to the attack of host antimicrobial peptides has been instrumental in eukaryogenesis and in the emergence of photosynthetic eukaryotes. Keywords: Chlamydomonas; targeting peptides; antimicrobial peptides; primary endosymbiosis; import into organelles; chloroplast; mitochondrion 1. -

The Photosynthetic Endosymbiont in Cryptomonad Cells Produces Both Chloroplast and Cytoplasmic-Type Ribosomes
Journal of Cell Science 107, 649-657 (1994) 649 Printed in Great Britain © The Company of Biologists Limited 1994 JCS6601 The photosynthetic endosymbiont in cryptomonad cells produces both chloroplast and cytoplasmic-type ribosomes Geoffrey I. McFadden1,*, Paul R. Gilson1 and Susan E. Douglas2 1Plant Cell Biology Research Centre, School of Botany, University of Melbourne, Parkville, Victoria, 3052, Australia 2Institute of Marine Biosciences, National Research Council of Canada, 1411 Oxford St, Halifax, Nova Scotia B3H 3Z1, Canada *Author for correspondence SUMMARY Cryptomonad algae contain a photosynthetic, eukaryotic tion machinery. We also localized transcripts of the host endosymbiont. The endosymbiont is much reduced but nucleus rRNA gene. These transcripts were found in the retains a small nucleus. DNA from this endosymbiont nucleolus of the host nucleus, and throughout the host nucleus encodes rRNAs, and it is presumed that these cytoplasm, but never in the endosymbiont compartment. rRNAs are incorporated into ribosomes. Surrounding the Our rRNA localizations indicate that the cryptomonad cell endosymbiont nucleus is a small volume of cytoplasm produces two different of sets of cytoplasmic-type proposed to be the vestigial cytoplasm of the endosymbiont. ribosomes in two separate subcellular compartments. The If this compartment is indeed the endosymbiont’s results suggest that there is no exchange of rRNAs between cytoplasm, it would be expected to contain ribosomes with these compartments. We also used the probe specific for the components encoded by the endosymbiont nucleus. In this endosymbiont rRNA gene to identify chromosomes from paper, we used in situ hybridization to localize rRNAs the endosymbiont nucleus in pulsed field gel electrophore- encoded by the endosymbiont nucleus of the cryptomonad sis. -
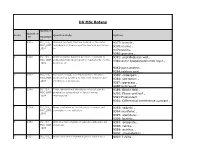
DU Msc Botany
DU MSc Botany Question Question Sr.No Question Body Options Id Descripti on 1 2345 DU_J19_ Channel proteins that are located in the outer 9377: barrels , MSC_BOT membrane of Gram-negative bacteria are known 9378:murins , _Q01 as 9379:porins, 9380:granules , 2 2346 DU_J19_ Gram-negative bacteria are more resistant to 9381: peptidoglycan wall., MSC_BOT antibiotics than Gram-positive bacteria due to the 9382:outer lipopolysaccharide layer., _Q02 presence of 9383:porin protein., 9384:teichoic acid. , 3 2347 DU_J19_ Dormant, tough, non-reproductive structure, 9385: endospore , MSC_BOT produced by bacteria to tide over unfavourable 9386: sclerotium , _Q03 conditions is known as: 9387:sporocarp , 9388:heterocyst , 4 2348 DU_J19_ Three-dimensional structures of a cell can be 9389: Bright field , MSC_BOT studied by using which of the following 9390: Phase contrast , _Q04 microscopes? 9391:Fluorescent , 9392: Differential interference contrast , 5 2349 DU_J19_ Algae that grow at the interface of water and 9393: epipelic , MSC_BOT atmosphere are called as 9394:neustonic , _Q05 9395: planktonic , 9396: benthic, 6 2350 DU_J19_ Orthorhombic crystals of calcium carbonate are 9397: aragonite., MSC_BOT known as 9398: calcite. , _Q06 9399: detritus. , 9400: stromatolites. , 7 2351 DU_J19_ Which one of the following amino acids has a 9401: Lysine , MSC_BOT nonpolar, aliphatic R group? _Q07 7 2351 DU_J19_ Which one of the following amino acids has a MSC_BOT nonpolar, aliphatic R group? 9402: Histidine , _Q07 9403: Arginine , 9404: Glycine, 8 2352 DU_J19_ -

Identification and Characterisation of Endogenous Avian Leukosis Virus Subgroup E (ALVE) Insertions in Chicken Whole Genome Sequencing Data Andrew S
Mason et al. Mobile DNA (2020) 11:22 https://doi.org/10.1186/s13100-020-00216-w RESEARCH Open Access Identification and characterisation of endogenous Avian Leukosis Virus subgroup E (ALVE) insertions in chicken whole genome sequencing data Andrew S. Mason1,2* , Ashlee R. Lund3, Paul M. Hocking1ˆ, Janet E. Fulton3† and David W. Burt1,4† Abstract Background: Endogenous retroviruses (ERVs) are the remnants of retroviral infections which can elicit prolonged genomic and immunological stress on their host organism. In chickens, endogenous Avian Leukosis Virus subgroup E (ALVE) expression has been associated with reductions in muscle growth rate and egg production, as well as providing the potential for novel recombinant viruses. However, ALVEs can remain in commercial stock due to their incomplete identification and association with desirable traits, such as ALVE21 and slow feathering. The availability of whole genome sequencing (WGS) data facilitates high-throughput identification and characterisation of these retroviral remnants. Results: We have developed obsERVer, a new bioinformatic ERV identification pipeline which can identify ALVEs in WGS data without further sequencing. With this pipeline, 20 ALVEs were identified across eight elite layer lines from Hy-Line International, including four novel integrations and characterisation of a fast feathered phenotypic revertant that still contained ALVE21. These bioinformatically detected sites were subsequently validated using new high- throughput KASP assays, which showed that obsERVer was highly precise and exhibited a 0% false discovery rate. A further fifty-seven diverse chicken WGS datasets were analysed for their ALVE content, identifying a total of 322 integration sites, over 80% of which were novel. Like exogenous ALV, ALVEs show site preference for proximity to protein-coding genes, but also exhibit signs of selection against deleterious integrations within genes. -

Can We Understand Evolution Without Symbiogenesis?
Can We Understand Evolution Without Symbiogenesis? Francisco Carrapiço …symbiosis is more than a mere casual and isolated biological phenomenon: it is in reality the most fundamental and universal order or law of life. Hermann Reinheimer (1915) Abstract This work is a contribution to the literature and knowledge on evolu- tion that takes into account the biological data obtained on symbiosis and sym- biogenesis. Evolution is traditionally considered a gradual process essentially consisting of natural selection, conducted on minimal phenotypical variations that are the result of mutations and genetic recombinations to form new spe- cies. However, the biological world presents and involves symbiotic associations between different organisms to form consortia, a new structural life dimension and a symbiont-induced speciation. The acknowledgment of this reality implies a new understanding of the natural world, in which symbiogenesis plays an important role as an evolutive mechanism. Within this understanding, symbiosis is the key to the acquisition of new genomes and new metabolic capacities, driving living forms’ evolution and the establishment of biodiversity and complexity on Earth. This chapter provides information on some of the key figures and their major works on symbiosis and symbiogenesis and reinforces the importance of these concepts in our understanding of the natural world and the role they play in the establishing of the evolutionary complexity of living systems. In this context, the concept of the symbiogenic superorganism is also discussed. Keywords Evolution · Symbiogenesis · Symbiosis · Symbiogenic superorgan- ism · New paradigm F. Carrapiço (*) Centre for Ecology Evolution and Environmental Change (CE3C); Centre for Philosophy of Science, Department of Plant Biology, Faculty of science, University of Lisbon, Lisbon, Portugal e-mail: [email protected] © Springer International Publishing Switzerland 2015 81 N. -

The Origin of Plastids By: Cheong Xin Chan, Ph.D
A Collaborative Learning Space for Science ABOUT FACULTY STUDENTS INTERMEDIATE CELL ORIGINS AND METABOLISM | Lead Editor: Gary Cote, Mario De Tullio The Origin of Plastids By: Cheong Xin Chan, Ph.D. & Debashish Bhattacharya, Ph.D. (Rutgers, The State University of New Jersey) © 2010 Nature Education Citation: Chan, C. X. & Bhattacharya, D. (2010) The Origin of Plastids. Nature Education 3(9):84 Plastids are core components of photosynthesis in plants and algae. Scientists are currently debating the events leading to the appearance of plastids in eukaryotic cells. Organelles, called plastids, are the main sites of photosynthesis in eukaryotic cells. Chloroplasts, as well as any other pigment containing cytoplasmic organelles that enables the harvesting and conversion of light and carbon dioxide into food and energy, are plastids. Found mainly in eukaryotic cells, plastids can be grouped into two distinctive types depending on their membrane structure: primary plastids and secondary plastids. Primary plastids are found in most algae and plants, and secondary, more-complex plastids are typically found in plankton, such as diatoms and dinoflagellates. Exploring the origin of plastids is an exciting field of research because it enhances our understanding of the basis of photosynthesis in green plants, our primary food source on planet Earth. Primary Plastids and Endosymbiosis Where did plastids originate? Their origin is explained by endosymbiosis, the act of a unicellular heterotrophic protist engulfing a free-living photosynthetic cyanobacterium and retaining it, instead of digesting it in the food vacuole (Margulis 1970; McFadden 2001; Kutschera & Niklas 2005). The captured cell (the endosymbiont) was then reduced to a functional organelle bound by two membranes, and was transmitted vertically to subsequent generations. -

Lynn Margulis
BIOGRAPHY THE INDOMITABLE EVOLUTIONIST: LYNN MARGULIS MEENAKSHI PANT The evolutionary “To me, the human move to take responsibility interest in science. In 1957, she graduated biologist Lynn for the living Earth is laughable — the rhetoric with a degree in Liberal Arts, and moved to Margulis is best of the powerless. The planet takes care of us, the University of Wisconsin to study Biology known for her not we of it. Our self-inflated moral imperative under Walter Plaut (who was to become her work on the Serial to guide a wayward Earth, or heal our sick supervisor) and Hans Ris. In 1960, she graduated Endosymbiotic planet, is evidence of our immense capacity with an MS degree in Zoology and Genetics. Theory (SET) to for self-delusion. Rather, we need to protect She then began her research career in the explain the origins ourselves from ourselves.” University of California, under the guidance of of eukaryotic Max Alfert, earning her doctoral degree in 1965. cells. This article Lynn was offered her first job — a research presents key facets his very courageous statement that assistantship and the position of a lecturer in in the life of Tchallenges the self-acclaimed supremacy Brandeis University — even before she could this avant-garde of humans over nature was made by Lynn finish her dissertation. However, it was only biologist and her Margulis (refer Fig.1). She is believed to be work that has one of the most creative scientific theorists after she had been awarded a PhD that she changed the way of the modern era, who transformed the idea moved to Boston University, where she taught we perceive life on of how life evolved on Earth. -

Restriction-Modification Systems As Mobile Genetic Elements in the Evolution of an Intracellular Symbiont
Restriction-Modification Systems as Mobile Genetic Elements in the Evolution of an Intracellular Symbiont Hao Zheng,y,1 Carsten Dietrich,y,1 Yuichi Hongoh,2 and Andreas Brune*,1 1Department of Biogeochemistry, Max Planck Institute for Terrestrial Microbiology, Marburg, Germany 2Department of Biological Sciences, Tokyo Institute of Technology, Tokyo, Japan yThese authors contributed equally to this work. *Corresponding author: E-mail: [email protected]. Associate editor: Eduardo Rocha Abstract Long-term vertical transmission of intracellular bacteria causes massive genomic erosion and results in extremely small genomes, particularly in ancient symbionts. Genome reduction is typically preceded by the accumulation of pseudogenes and proliferation of mobile genetic elements, which are responsible for chromosome rearrangements during the initial stage of endosymbiosis. We compared the genomes of an endosymbiont of termite gut flagellates, “Candidatus Endomicrobium trichonymphae,” and its free-living relative Endomicrobium proavitum anddiscoveredmanyremnants of restriction-modification (R-M) systems that are consistently associated with genome rearrangements in the endosym- biont genome. The rearrangements include apparent insertions, transpositions, and the duplication of a genomic region; there was no evidence of transposon structures or other mobile elements. Our study reveals a so far unrecognized mechanism for genome rearrangements in intracellular symbionts and sheds new light on the general role of R-M systems in genome evolution. Key words: restriction-modification system, CRISPR-Cas system, mobile genetic elements, endosymbiont, genome reduction. Mutualistic symbioses between bacteria and eukaryotes are indicate that it is still in an early stage of genome reduction ubiquitous in nature, and they can greatly affect the genomes (Hongoh et al. 2008). -

Cooperative Evolution Reclaiming Darwin’S Vision
COOPERATIVE EVOLUTION RECLAIMING DARWIN’S VISION COOPERATIVE EVOLUTION RECLAIMING DARWIN’S VISION CHRISTOPHER BRYANT AND VALERIE A. BROWN For Annie Bryant who has had to put up with parasites and mitochondria at the dinner table for 58 years and, in spite of that, managed to pass on to our children a very healthy mitochondrial genome, with love. And for Chris, Sarah, Elliot and Amon Brown who are taking the next steps in cooperative evolution. By mutual confidence and mutual aid, Great deeds are done and great discoveries made. – Homer Published by ANU Press The Australian National University Acton ACT 2601, Australia Email: [email protected] Available to download for free at press.anu.edu.au ISBN (print): 9781760464288 ISBN (online): 9781760464295 WorldCat (print): 1240754622 WorldCat (online): 1240754606 DOI: 10.22459/CE.2021 This title is published under a Creative Commons Attribution-NonCommercial- NoDerivatives 4.0 International (CC BY-NC-ND 4.0). The full licence terms are available at creativecommons.org/licenses/by-nc-nd/4.0/legalcode Cover design and layout by ANU Press This edition © 2021 ANU Press CONTENTS Acknowledgements . xi Glossary of Words and Phrases . xiii Introduction . 1 1 . In Homage to Darwin . 5 2 . All Knowledge is Metaphor . 17 3 . Intelligent Evolution and Intelligence . 31 4 . How Evolution Works . 55 5 . The Past is a Foreign Country . 75 6 . We Do Things Differently Now . 89 7 . Energy: Where it all Begins . 103 8 . Everything is Connected . 119 9 . Walling In and Walling Out . 133 10 . Becoming Human . 149 11 . Inheriting the Earth . 163 12 . Our Closest Cousins .