Relevance of the COPI Complex for Alzheimer's Disease Progression In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

Transcriptomic Analysis of the Aquaporin (AQP) Gene Family
Pancreatology 19 (2019) 436e442 Contents lists available at ScienceDirect Pancreatology journal homepage: www.elsevier.com/locate/pan Transcriptomic analysis of the Aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma Dimitrios E. Magouliotis a, b, Vasiliki S. Tasiopoulou c, Konstantinos Dimas d, * Nikos Sakellaridis d, Konstantina A. Svokos e, Alexis A. Svokos f, Dimitris Zacharoulis b, a Division of Surgery and Interventional Science, Faculty of Medical Sciences, UCL, London, UK b Department of Surgery, University of Thessaly, Biopolis, Larissa, Greece c Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece d Department of Pharmacology, Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece e The Warren Alpert Medical School of Brown University, Providence, RI, USA f Riverside Regional Medical Center, Newport News, VA, USA article info abstract Article history: Background: This study aimed to assess the differential gene expression of aquaporin (AQP) gene family Received 14 October 2018 interactome in pancreatic ductal adenocarcinoma (PDAC) using data mining techniques to identify novel Received in revised form candidate genes intervening in the pathogenicity of PDAC. 29 January 2019 Method: Transcriptome data mining techniques were used in order to construct the interactome of the Accepted 9 February 2019 AQP gene family and to determine which genes members are differentially expressed in PDAC as Available online 11 February 2019 compared to controls. The same techniques were used in order to evaluate the potential prognostic role of the differentially expressed genes. Keywords: PDAC Results: Transcriptome microarray data of four GEO datasets were incorporated, including 142 primary Aquaporin tumor samples and 104 normal pancreatic tissue samples. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
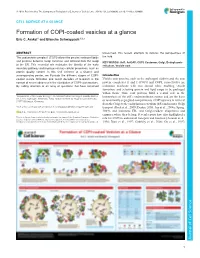
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong. -

Overlapping Role of SCYL1 and SCYL3 in Maintaining Motor Neuron Viability
The Journal of Neuroscience, March 7, 2018 • 38(10):2615–2630 • 2615 Neurobiology of Disease Overlapping Role of SCYL1 and SCYL3 in Maintaining Motor Neuron Viability Emin Kuliyev,1 Sebastien Gingras,4 XClifford S. Guy,1 Sherie Howell,2 Peter Vogel,3 and XStephane Pelletier1 1Departments of Immunology, 2Pathology, 3Veterinary Pathology Core, Advanced Histology Core, St. Jude Children’s Research Hospital, Memphis, Tennessee 38105, and 4Department of Immunology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15213 Members of the SCY1-like (SCYL) family of protein kinases are evolutionarily conserved and ubiquitously expressed proteins character- ized by an N-terminal pseudokinase domain, centrally located Huntingtin, elongation factor 3, protein phosphatase 2A, yeast kinase TOR1 repeats, and an overall disorganized C-terminal segment. In mammals, three family members encoded by genes Scyl1, Scyl2, and Scyl3 have been described. Studies have pointed to a role for SCYL1 and SCYL2 in regulating neuronal function and viability in mice and humans, but little is known about the biological function of SCYL3. Here, we show that the biochemical and cell biological properties of SCYL3aresimilartothoseofSCYL1andbothproteinsworkinconjunctiontomaintainmotorneuronviability.Specifically,althoughlack of Scyl3 in mice has no apparent effect on embryogenesis and postnatal life, it accelerates the onset of the motor neuron disorder caused by Scyl1 deficiency. Growth abnormalities, motor dysfunction, hindlimb paralysis, muscle wasting, neurogenic atrophy, motor neuron degeneration, and loss of large-caliber axons in peripheral nerves occurred at an earlier age in Scyl1/Scyl3 double-deficient mice than in Scyl1-deficientmice.DiseaseonsetalsocorrelatedwiththemislocalizationofTDP-43inspinalmotorneurons,suggestingthatSCYL1and SCYL3 regulate TDP-43 proteostasis. Together, our results demonstrate an overlapping role for SCYL1 and SCYL3 in vivo and highlight the importance the SCYL family of proteins in regulating neuronal function and survival. -

Dissertation
DISSERTATION submitted to the Combined Faculty of Natural Sciences and Mathematics of the Ruperto Carola University Heidelberg, Germany for the degree of Doctor of Natural Sciences Presented by Manu Jain Goyal (M.Sc, Biotechnology) born in Kota, India Oral examination: 03.07.2019 Paralog specific role of COPI pathway in P19 neuronal differentiation Referees: Prof. Dr. Felix Wieland Dr. Julien Béthune This work has been carried out at the Biochemistry Center of the University of Heidelberg from January 2015 to January 2019 under the supervision of Dr. Julien Béthune. Acknowledgement I would like to thank Julien for giving me opportunity to be part of his team, for supervising me throughout my PhD. I appreciate that all the scientific discussions we have had helped me to improve my knowledge and thinking scientifically. Thank you so much for giving freedom in the lab to work independently and for handling the situations in a best possible way. Next, I would like to thank my TAC members Prof. Dr. Felix and Prof. Dr. Walter for giving suggestions and inputs for my project. Also providing me antibodies and equipments for my experiments. I would like to thank my examiners Prof Dr. Thomas Söllner and Prof. Dr. Marius Lemberg for considering it. I would like to thank all my team members and special thanks to Petra, Cinthia and Isabel for always supporting and helping me. Additionally, I would like to thank Alice and Laura for socializing together outside the BZH so that I can survive far away from my home country. I admire all the moments I had with you all. -

Consequences of Mitotic Loss of Heterozygosity on Genomic Imprinting in Mouse Embryonic Stem Cells
CONSEQUENCES OF MITOTIC LOSS OF HETEROZYGOSITY ON GENOMIC IMPRINTING IN MOUSE EMBRYONIC STEM CELLS by RACHEL LEIGH ELVES B.Sc., University of British Columbia, 2004 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIRMENTS FOR THE DEGREE OF MASTER OF SCIENCE in THE FACULTY OF GRADUATE STUDIES (Medical Genetics) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) August 2008 © Rachel Leigh Elves, 2008 Abstract Epigenetic differences between maternally inherited and paternally inherited chromosomes, such as CpG methylation, render the maternal and paternal genome functionally inequivalent, a phenomenon called genomic imprinting. This functional inequivalence is exemplified with imprinted genes, whose expression is parent-of-origin specific. The dosage of imprinted gene expression is disrupted in cells with uniparental disomy (UPD), which is an unequal parental contribution to the genome. I have derived mouse embryonic stem (ES) cell sub-lines with maternal UPD (mUPD) for mouse chromosome 6 (MMU6) to characterize regulation and maintenance of imprinted gene expression. The main finding from this study is that maintenance of imprinting in mitotic UPD is extremely variable. Imprint maintenance was shown to vary from gene to gene, and to vary between ES cell lines depending on the mechanism of loss of heterozygosity (LOH) in that cell line. Certain genes analyzed, such as Peg10 , Sgce , Peg1 , and Mit1 showed abnormal expression in ES cell lines for which they were mUPD. These abnormal expression levels are similar to that observed in ES cells with meiotically-derived full genome mUPD (parthenogenetic ES cells). Imprinted CpG methylation at the Peg1 promoter was found to be abnormal in all sub-lines with mUPD for Peg1 . -

Supplementary Table S2
1-high in cerebrotropic Gene P-value patients Definition BCHE 2.00E-04 1 Butyrylcholinesterase PLCB2 2.00E-04 -1 Phospholipase C, beta 2 SF3B1 2.00E-04 -1 Splicing factor 3b, subunit 1 BCHE 0.00022 1 Butyrylcholinesterase ZNF721 0.00028 -1 Zinc finger protein 721 GNAI1 0.00044 1 Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 GNAI1 0.00049 1 Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 PDE1B 0.00069 -1 Phosphodiesterase 1B, calmodulin-dependent MCOLN2 0.00085 -1 Mucolipin 2 PGCP 0.00116 1 Plasma glutamate carboxypeptidase TMX4 0.00116 1 Thioredoxin-related transmembrane protein 4 C10orf11 0.00142 1 Chromosome 10 open reading frame 11 TRIM14 0.00156 -1 Tripartite motif-containing 14 APOBEC3D 0.00173 -1 Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D ANXA6 0.00185 -1 Annexin A6 NOS3 0.00209 -1 Nitric oxide synthase 3 SELI 0.00209 -1 Selenoprotein I NYNRIN 0.0023 -1 NYN domain and retroviral integrase containing ANKFY1 0.00253 -1 Ankyrin repeat and FYVE domain containing 1 APOBEC3F 0.00278 -1 Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F EBI2 0.00278 -1 Epstein-Barr virus induced gene 2 ETHE1 0.00278 1 Ethylmalonic encephalopathy 1 PDE7A 0.00278 -1 Phosphodiesterase 7A HLA-DOA 0.00305 -1 Major histocompatibility complex, class II, DO alpha SOX13 0.00305 1 SRY (sex determining region Y)-box 13 ABHD2 3.34E-03 1 Abhydrolase domain containing 2 MOCS2 0.00334 1 Molybdenum cofactor synthesis 2 TTLL6 0.00365 -1 Tubulin tyrosine ligase-like family, member 6 SHANK3 0.00394 -1 SH3 and multiple ankyrin repeat domains 3 ADCY4 0.004 -1 Adenylate cyclase 4 CD3D 0.004 -1 CD3d molecule, delta (CD3-TCR complex) (CD3D), transcript variant 1, mRNA. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
Supplemental Figure 2.Eps
Androgen Resveratrol 75uM 150uM 43h-10nM 43h-100nM 43h-1uM 20h-10uM 18h-25uM 20h-40uM 20h-100uM 0h 1h 3h 6h 9h 12h 15h 18h 21h 24h 48h 60h 0h 1h 2h 3h 4h 6h 8h 12h 14h 16h 18h 20h 22h 24h 48h 60h 7h-R1881 9h-R1881 18h-R1881 24h-R1881 50h-R1881 72h-R1881 18h-DHT10 50h-DHT10 24h-DHT100 24h-DHT1000 46h-noR1881 70h-noR1881 99855 ZFH4 zinc finger homeodomain 4 Hs.109314 105366 NFKB1 nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 p105 Hs.83428 112015 SEPT6 septin 6 Hs.90998 111438 SLC12A2 **solute carrier family 12 sodium/potassium/chloride transporters, member 2 Hs.110736 117460 ZIC2 Zic family member 2 odd-paired homolog, Drosophila Hs.132863 112768 KIAA0914 KIAA0914 gene product Hs.177664 111066 ESTs, Weakly similar to GNMSLL retrovirus-related reverse transcriptase homolog - mouse retrotransposon [M.musculus] Hs.301451 104992 FN1 fibronectin 1 Hs.287820 116170 RDC1 G protein-coupled receptor Hs.23016 99709 PDCD5 programmed cell death 5 Hs.166468 112624 CTBP1 C-terminal binding protein 1 Hs.239737 118259 GRB10 growth factor receptor-bound protein 10 Hs.81875 111378 PURA purine-rich element binding protein A Hs.29117 120755 **ESTs, Weakly similar to CNG1_HUMAN cGMP-gated cation channel alpha 1 CNG channel alpha 1 CNG-1 CNG1 Cyclic nucleotide gated channel alpha 1 Cyclic nucleotide gated channel, photoreceptor Hs.356525 118729 DKFZP564D0462 hypothetical protein DKFZp564D0462 Hs.44197 101077 LRBA vesicle trafficking, beach and anchor containing Hs.62354 116608 BTD biotinidase Hs.78885 107627 BCHE butyrylcholinesterase -

Disease-Related Cellular Protein Networks Differentially Affected
www.nature.com/scientificreports OPEN Disease‑related cellular protein networks diferentially afected under diferent EGFR mutations in lung adenocarcinoma Toshihide Nishimura1,8*, Haruhiko Nakamura1,2,8, Ayako Yachie3,8, Takeshi Hase3,8, Kiyonaga Fujii1,8, Hirotaka Koizumi4, Saeko Naruki4, Masayuki Takagi4, Yukiko Matsuoka3, Naoki Furuya5, Harubumi Kato6,7 & Hisashi Saji2 It is unclear how epidermal growth factor receptor EGFR major driver mutations (L858R or Ex19del) afect downstream molecular networks and pathways. This study aimed to provide information on the infuences of these mutations. The study assessed 36 protein expression profles of lung adenocarcinoma (Ex19del, nine; L858R, nine; no Ex19del/L858R, 18). Weighted gene co-expression network analysis together with analysis of variance-based screening identifed 13 co-expressed modules and their eigen proteins. Pathway enrichment analysis for the Ex19del mutation demonstrated involvement of SUMOylation, epithelial and mesenchymal transition, ERK/mitogen- activated protein kinase signalling via phosphorylation and Hippo signalling. Additionally, analysis for the L858R mutation identifed various pathways related to cancer cell survival and death. With regard to the Ex19del mutation, ROCK, RPS6KA1, ARF1, IL2RA and several ErbB pathways were upregulated, whereas AURK and GSKIP were downregulated. With regard to the L858R mutation, RB1, TSC22D3 and DOCK1 were downregulated, whereas various networks, including VEGFA, were moderately upregulated. In all mutation types, CD80/CD86 (B7), MHC, CIITA and IFGN were activated, whereas CD37 and SAFB were inhibited. Costimulatory immune-checkpoint pathways by B7/CD28 were mainly activated, whereas those by PD-1/PD-L1 were inhibited. Our fndings may help identify potential therapeutic targets and develop therapeutic strategies to improve patient outcomes. -

An Integrated Approach to Comprehensively Map the Molecular Context of Proteins
bioRxiv preprint doi: https://doi.org/10.1101/264788; this version posted February 13, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Liu et al. An integrated approach to comprehensively map the molecular context of proteins Xiaonan Liu1,2, Kari Salokas1,2, Fitsum Tamene1,2,3 and Markku Varjosalo1,2,3* 1Institute of Biotechnology, University of Helsinki, Helsinki 00014, Finland 2Helsinki Institute of Life Science, University of Helsinki, Helsinki 00014, Finland 3Proteomics Unit, University of Helsinki, Helsinki 00014, Finland *Corresponding author Abstract: Protein-protein interactions underlie almost all cellular functions. The comprehensive mapping of these complex cellular networks of stable and transient associations has been made available by affi nity purifi cation mass spectrometry (AP-MS) and more recently by proximity based labelling methods such as BioID. Due the advancements in both methods and MS instrumentation, an in-depth analysis of the whole human proteome is at grasps. In order to facilitate this, we designed and optimized an integrated approach utilizing MAC-tag combining both AP-MS and BioID in a single construct. We systematically applied this approach to 18 subcellular localization markers and generated a molecular context database, which can be used to defi ne molecular locations for any protein of interest. In addition, we show that by combining the AP-MS and BioID results we can also obtain interaction distances within a complex. Taken together, our combined strategy off ers comprehensive approach for mapping physical and functional protein interactions. Introduction: Majority of proteins do not function in isolation geting the endogenous bait protein, allowing and their interactions with other proteins defi ne purifi cation of the bait protein together with the their cellular functions.