Parallel Host Range Expansion in Two Unrelated Cossid Moths Infesting
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Butterflies of Ontario & Summaries of Lepidoptera
ISBN #: 0-921631-12-X BUTTERFLIES OF ONTARIO & SUMMARIES OF LEPIDOPTERA ENCOUNTERED IN ONTARIO IN 1991 BY A.J. HANKS &Q.F. HESS PRODUCTION BY ALAN J. HANKS APRIL 1992 CONTENTS 1. INTRODUCTION PAGE 1 2. WEATHER DURING THE 1991 SEASON 6 3. CORRECTIONS TO PREVIOUS T.E.A. SUMMARIES 7 4. SPECIAL NOTES ON ONTARIO LEPIDOPTERA 8 4.1 The Inornate Ringlet in Middlesex & Lambton Cos. 8 4.2 The Monarch in Ontario 8 4.3 The Status of the Karner Blue & Frosted Elfin in Ontario in 1991 11 4.4 The West Virginia White in Ontario in 1991 11 4.5 Butterfly & Moth Records for Kettle Point 11 4.6 Butterflies in the Hamilton Study Area 12 4.7 Notes & Observations on the Early Hairstreak 15 4.8 A Big Day for Migrants 16 4.9 The Ocola Skipper - New to Ontario & Canada .17 4.10 The Brazilian Skipper - New to Ontario & Canada 19 4.11 Further Notes on the Zarucco Dusky Wing in Ontario 21 4.12 A Range Extension for the Large Marblewing 22 4.13 The Grayling North of Lake Superior 22 4.14 Description of an Aberrant Crescent 23 4.15 A New Foodplant for the Old World Swallowtail 24 4.16 An Owl Moth at Point Pelee 25 4.17 Butterfly Sampling in Algoma District 26 4.18 Record Early Butterfly Dates in 1991 26 4.19 Rearing Notes from Northumberland County 28 5. GENERAL SUMMARY 29 6. 1990 SUMMARY OF ONTARIO BUTTERFLIES, SKIPPERS & MOTHS 32 Hesperiidae 32 Papilionidae 42 Pieridae 44 Lycaenidae 48 Libytheidae 56 Nymphalidae 56 Apaturidae 66 Satyr1dae 66 Danaidae 70 MOTHS 72 CONTINUOUS MOTH CYCLICAL SUMMARY 85 7. -

Biosecurity Plan for the Vegetable Industry
Biosecurity Plan for the Vegetable Industry A shared responsibility between government and industry Version 3.0 May 2018 Plant Health AUSTRALIA Location: Level 1 1 Phipps Close DEAKIN ACT 2600 Phone: +61 2 6215 7700 Fax: +61 2 6260 4321 E-mail: [email protected] Visit our web site: www.planthealthaustralia.com.au An electronic copy of this plan is available through the email address listed above. © Plant Health Australia Limited 2018 Copyright in this publication is owned by Plant Health Australia Limited, except when content has been provided by other contributors, in which case copyright may be owned by another person. With the exception of any material protected by a trade mark, this publication is licensed under a Creative Commons Attribution-No Derivs 3.0 Australia licence. Any use of this publication, other than as authorised under this licence or copyright law, is prohibited. http://creativecommons.org/licenses/by-nd/3.0/ - This details the relevant licence conditions, including the full legal code. This licence allows for redistribution, commercial and non-commercial, as long as it is passed along unchanged and in whole, with credit to Plant Health Australia (as below). In referencing this document, the preferred citation is: Plant Health Australia Ltd (2018) Biosecurity Plan for the Vegetable Industry (Version 3.0 – 2018) Plant Health Australia, Canberra, ACT. This project has been funded by Hort Innovation, using the vegetable research and development levy and contributions from the Australian Government. Hort Innovation is the grower-owned, not for profit research and development corporation for Australian horticulture Disclaimer: The material contained in this publication is produced for general information only. -

Lepidoptera of North America 5
Lepidoptera of North America 5. Contributions to the Knowledge of Southern West Virginia Lepidoptera Contributions of the C.P. Gillette Museum of Arthropod Diversity Colorado State University Lepidoptera of North America 5. Contributions to the Knowledge of Southern West Virginia Lepidoptera by Valerio Albu, 1411 E. Sweetbriar Drive Fresno, CA 93720 and Eric Metzler, 1241 Kildale Square North Columbus, OH 43229 April 30, 2004 Contributions of the C.P. Gillette Museum of Arthropod Diversity Colorado State University Cover illustration: Blueberry Sphinx (Paonias astylus (Drury)], an eastern endemic. Photo by Valeriu Albu. ISBN 1084-8819 This publication and others in the series may be ordered from the C.P. Gillette Museum of Arthropod Diversity, Department of Bioagricultural Sciences and Pest Management Colorado State University, Fort Collins, CO 80523 Abstract A list of 1531 species ofLepidoptera is presented, collected over 15 years (1988 to 2002), in eleven southern West Virginia counties. A variety of collecting methods was used, including netting, light attracting, light trapping and pheromone trapping. The specimens were identified by the currently available pictorial sources and determination keys. Many were also sent to specialists for confirmation or identification. The majority of the data was from Kanawha County, reflecting the area of more intensive sampling effort by the senior author. This imbalance of data between Kanawha County and other counties should even out with further sampling of the area. Key Words: Appalachian Mountains, -

Making a Meeting Place Eucalypt Trail Map and Signs
Friends of the Australian National Botanic Gardens Number 73 April 2013 Inside: Making a Meeting Place Eucalypt Trail map and signs Friends of the Australian National Botanic Gardens Patron His Excellency Mr Michael Bryce AM AE Vice Patron Mrs Marlena Jeffery President David Coutts Vice President Barbara Podger Secretary John Connolly Treasurer Marion Jones Public Officer David Coutts General Committee Dennis Ayliffe Glenys Bishop Anne Campbell Lesley Jackman Warwick Wright Talks Convenor Lesley Jackman Membership Secretary Barbara Scott Fronds Committee Margaret Clarke Barbara Podger Anne Rawson Growing Friends Kath Holtzapffel Botanic Art Groups Helen Hinton Photographic Group Graham Brown Social events Jan Finley Exec. Director, ANBG Dr Judy West Post: Friends of ANBG, GPO Box 1777 Canberra ACT 2601 Australia Telephone: (02) 6250 9548 (messages) Internet: www.friendsanbg.org.au Email addresses: [email protected] IN THIS ISSUE [email protected] [email protected] Eucalypt Trail map and signs.................................................2 Fronds is published three times a year. We welcome your articles for inclusion in the next Discover eucalypts ................................................................3 issue. Material should be forwarded to the Giant wood moths in the Gardens .........................................4 Fronds Committee by mid-February for the April issue; mid-June for the August issue; Growing Friends ....................................................................6 mid-October -

Animal and Plant Health Inspection Service, USDA § 319.56–2H
Animal and Plant Health Inspection Service, USDA § 319.56–2h pest conditions on arrival or to assure fect that such garlic is free of living himself of the effectiveness of the stages of Brachycerus spp. and treatment. Dyspessa ulula (Bkh.), said certifi- (c) The entry of cipollini from Mo- cation to be based on field inspection rocco may be made only through the and certification and subsequent reex- ports of New York and Boston at which amination at the port of departure ports facilities for vacuum fumigation prior to exportation. The with methyl bromide, as herein re- phytosanitary certificate to be issued quired, are available. by such official shall show the ship- [24 FR 10788, Dec. 29, 1959. Redesignated at 50 ment to be either initially free from FR 9788, Mar. 12, 1985] these pests or to have been fumigated. (ii) The original copy of the § 319.56–2f [Reserved] phytosanitary certificate shall be at- tached to the manifest accompanying § 319.56–2g Administrative instruc- the shipment. However, with the con- tions prescribing method of treat- sent of the Plant Quarantine inspector, ment of garlic from specified coun- tries. the importer may arrange to have the original phytosanitary certificate (a) Except as otherwise provided in mailed direct to the Inspector in these administrative instructions, fu- Charge, Plant Protection and Quar- migation with methyl bromide in vacu- antine Programs, at the port of entry, um fumigation chambers, in accord- if this will expedite inspection and re- ance with the Plant Protection and lease of certified shipments. If such an Quarantine Treatment Manual, which arrangement is made, a copy of the is incorporated by reference at § 300.1 of phytosanitary certificate shall be at- this chapter, is a condition of entry tached to the manifest accompanying under permit for all shipments of garlic the shipment. -

Alien Invasive Species and International Trade
Forest Research Institute Alien Invasive Species and International Trade Edited by Hugh Evans and Tomasz Oszako Warsaw 2007 Reviewers: Steve Woodward (University of Aberdeen, School of Biological Sciences, Scotland, UK) François Lefort (University of Applied Science in Lullier, Switzerland) © Copyright by Forest Research Institute, Warsaw 2007 ISBN 978-83-87647-64-3 Description of photographs on the covers: Alder decline in Poland – T. Oszako, Forest Research Institute, Poland ALB Brighton – Forest Research, UK; Anoplophora exit hole (example of wood packaging pathway) – R. Burgess, Forestry Commission, UK Cameraria adult Brussels – P. Roose, Belgium; Cameraria damage medium view – Forest Research, UK; other photographs description inside articles – see Belbahri et al. Language Editor: James Richards Layout: Gra¿yna Szujecka Print: Sowa–Print on Demand www.sowadruk.pl, phone: +48 022 431 81 40 Instytut Badawczy Leœnictwa 05-090 Raszyn, ul. Braci Leœnej 3, phone [+48 22] 715 06 16 e-mail: [email protected] CONTENTS Introduction .......................................6 Part I – EXTENDED ABSTRACTS Thomas Jung, Marla Downing, Markus Blaschke, Thomas Vernon Phytophthora root and collar rot of alders caused by the invasive Phytophthora alni: actual distribution, pathways, and modeled potential distribution in Bavaria ......................10 Tomasz Oszako, Leszek B. Orlikowski, Aleksandra Trzewik, Teresa Orlikowska Studies on the occurrence of Phytophthora ramorum in nurseries, forest stands and garden centers ..........................19 Lassaad Belbahri, Eduardo Moralejo, Gautier Calmin, François Lefort, Jose A. Garcia, Enrique Descals Reports of Phytophthora hedraiandra on Viburnum tinus and Rhododendron catawbiense in Spain ..................26 Leszek B. Orlikowski, Tomasz Oszako The influence of nursery-cultivated plants, as well as cereals, legumes and crucifers, on selected species of Phytophthopra ............30 Lassaad Belbahri, Gautier Calmin, Tomasz Oszako, Eduardo Moralejo, Jose A. -

Full Page Photo
Эверсманния. Энтомологические исследования Eversmannia в России и соседних регионах. Вып. 9. 20.III.2007: 11–33 No. 9. 2007. Р. В. Яковлев г. Барнаул, Алтайский государственный университет (Южно-Сибирский ботанический сад), Алтайский краевой институт повышения квалификации работников образования Древоточцы (Lepidoptera: Cossidae) России R.V. Yakovlev. Carpenter moths (Lepidoptera: Cossidae) of Russia. SUMMARY. In this article the catalogue of Cossidae of Russia is presented. An annotated list including 32 species has been compiled. Two new species and two new subspecies: Catopta albonubila argunica, ssp. n. (Locus typicus: East Trasbaicalia, Kuenga, 45 km SW. Sretensk), Catopta perunovi, sp. n. (Locus typicus: Russia, Altai Rep., near Ongudai), Cossus cossus dauricus, ssp. n. (Locus typicus: Russia Transbaikalia, Chita Province, Nizhnii Tsasutchei), Phragmataecia pacifica, sp. n. (Locus typicus: Russia, Dagestan, 5 km E. Urma) are described. Besides, a new species from S. Mongolia Catopta saldaitisi, sp. n. (Locus typicus: S. Mongolia, Omnogovi Aimak, the Gobi Altai Mts., Gurvan Sayhan) is described. For each species bibliography and distribution map are given. Проводимая в последние годы работа по исследованию древоточцев (Сossidae) Евразии при- вела к описанию ряда новых таксонов с территории Палеарктики, позволила установить грани- цы распространения многих видов. В данной работе делается попытка представить полный ка- талог древоточцев России, выполненный как на основании анализа литературных данных, так и на большом фактическом материале. Исследованный материал хранится в ряде коллекций Рос- сии, Украины, Германии, Австрии. Приведен аннотированный список видов, включающий дан- ные о первичном описании, синонимии, месте хранения типового материала и распростране- нии. Для редких и малоизвестных видов и для новых таксонов приводятся полные данные эти- кеток. Для каждого вида приведена карта распространения на территории России. -

Bioblitz! OK 2019 - Cherokee County Moth List
BioBlitz! OK 2019 - Cherokee County Moth List Sort Family Species 00366 Tineidae Acrolophus mortipennella 00372 Tineidae Acrolophus plumifrontella Eastern Grass Tubeworm Moth 00373 Tineidae Acrolophus popeanella 00383 Tineidae Acrolophus texanella 00457 Psychidae Thyridopteryx ephemeraeformis Evergreen Bagworm Moth 01011 Oecophoridae Antaeotricha schlaegeri Schlaeger's Fruitworm 01014 Oecophoridae Antaeotricha leucillana 02047 Gelechiidae Keiferia lycopersicella Tomato Pinworm 02204 Gelechiidae Fascista cercerisella 02301.2 Gelechiidae Dichomeris isa 02401 Yponomeutidae Atteva aurea 02401 Yponomeutidae Atteva aurea Ailanthus Webworm Moth 02583 Sesiidae Synanthedon exitiosa 02691 Cossidae Fania nanus 02694 Cossidae Prionoxystus macmurtrei Little Carpenterworm Moth 02837 Tortricidae Olethreutes astrologana The Astrologer 03172 Tortricidae Epiblema strenuana 03202 Tortricidae Epiblema otiosana 03494 Tortricidae Cydia latiferreanus Filbert Worm 03573 Tortricidae Decodes basiplaganus 03632 Tortricidae Choristoneura fractittana 03635 Tortricidae Choristoneura rosaceana Oblique-banded Leafroller moth 03688 Tortricidae Clepsis peritana 03695 Tortricidae Sparganothis sulfureana Sparganothis Fruitworm Moth 03732 Tortricidae Platynota flavedana 03768.99 Tortricidae Cochylis ringsi 04639 Zygaenidae Pyromorpha dimidiata Orange-patched Smoky Moth 04644 Megalopygidae Lagoa crispata Black Waved Flannel Moth 04647 Megalopygidae Megalopyge opercularis 04665 Limacodidae Lithacodes fasciola 04677 Limacodidae Phobetron pithecium Hag Moth 04691 Limacodidae -

The Entomofauna on Eucalyptus in Israel: a Review
EUROPEAN JOURNAL OF ENTOMOLOGYENTOMOLOGY ISSN (online): 1802-8829 Eur. J. Entomol. 116: 450–460, 2019 http://www.eje.cz doi: 10.14411/eje.2019.046 REVIEW The entomofauna on Eucalyptus in Israel: A review ZVI MENDEL and ALEX PROTASOV Department of Entomology, Institute of Plant Protection, Agricultural Research Organization, The Volcani Center, Rishon LeTzion 7528809, Israel; e-mails: [email protected], [email protected] Key words. Eucalyptus, Israel, invasive species, native species, insect pests, natural enemies Abstract. The fi rst successful Eucalyptus stands were planted in Israel in 1884. This tree genus, particularly E. camaldulensis, now covers approximately 11,000 ha and constitutes nearly 4% of all planted ornamental trees. Here we review and discuss the information available about indigenous and invasive species of insects that develop on Eucalyptus trees in Israel and the natural enemies of specifi c exotic insects of this tree. Sixty-two phytophagous species are recorded on this tree of which approximately 60% are indigenous. The largest group are the sap feeders, including both indigenous and invasive species, which are mostly found on irrigated trees, or in wetlands. The second largest group are wood feeders, polyphagous Coleoptera that form the dominant native group, developing in dying or dead wood. Most of the seventeen parasitoids associated with the ten invasive Eucalyptus-specifi c species were introduced as biocontrol agents in classical biological control projects. None of the polyphagous species recorded on Eucalyptus pose any threat to this tree. The most noxious invasive specifi c pests, the gall wasps (Eulophidae) and bronze bug (Thaumastocoris peregrinus), are well controlled by introduced parasitoids. -

Moths of the Douglas Lake Region (Emmet and Cheboygan Counties), Michigan: VI
The Great Lakes Entomologist Volume 35 Number 1 - Spring/Summer 2002 Number 1 - Article 10 Spring/Summer 2002 April 2002 Moths of the Douglas Lake Region (Emmet and Cheboygan Counties), Michigan: VI. Miscellaneous Small Families (Lepidoptera) Edward G. Voss University of Michigan Follow this and additional works at: https://scholar.valpo.edu/tgle Part of the Entomology Commons Recommended Citation Voss, Edward G. 2002. "Moths of the Douglas Lake Region (Emmet and Cheboygan Counties), Michigan: VI. Miscellaneous Small Families (Lepidoptera)," The Great Lakes Entomologist, vol 35 (1) Available at: https://scholar.valpo.edu/tgle/vol35/iss1/10 This Peer-Review Article is brought to you for free and open access by the Department of Biology at ValpoScholar. It has been accepted for inclusion in The Great Lakes Entomologist by an authorized administrator of ValpoScholar. For more information, please contact a ValpoScholar staff member at [email protected]. Voss: Moths of the Douglas Lake Region (Emmet and Cheboygan Counties), 2002 THE GREAT LAKES ENTOMOLOGIST 53 MOTHS OF THE DOUGLAS LAKE REGION (EMMET AND CHEBOYGAN COUNTIES), MICHIGAN: VI. MISCELLANEOUS SMALL FAMILIES (LEPIDOPTERA) Edward G. Voss1 ABSTRACT Forty-seven species in nine families of Lepidoptera (Hepialidae, Psychidae, Alucitidae, Sesiidae, Cossidae, Limacodidae, Thyrididae, Pterophoridae, Epiplemi- dae) are listed with earliest and latest recorded flight dates in Emmet and Cheboy- gan counties, which share the northern tip of the Lower Peninsula of Michigan. The records are from the principal institutional and private collections of Michigan moths and continue the documented listing of Lepidoptera in the region. ____________________ Emmet and Cheboygan counties share the northern tip of the Lower Peninsula of Michigan, the former bordered on the west by Lake Michigan and the latter, on the east by Lake Huron. -
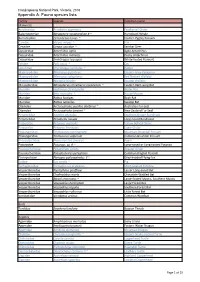
Report-VIC-Croajingolong National Park-Appendix A
Croajingolong National Park, Victoria, 2016 Appendix A: Fauna species lists Family Species Common name Mammals Acrobatidae Acrobates pygmaeus Feathertail Glider Balaenopteriae Megaptera novaeangliae # ~ Humpback Whale Burramyidae Cercartetus nanus ~ Eastern Pygmy Possum Canidae Vulpes vulpes ^ Fox Cervidae Cervus unicolor ^ Sambar Deer Dasyuridae Antechinus agilis Agile Antechinus Dasyuridae Antechinus mimetes Dusky Antechinus Dasyuridae Sminthopsis leucopus White-footed Dunnart Felidae Felis catus ^ Cat Leporidae Oryctolagus cuniculus ^ Rabbit Macropodidae Macropus giganteus Eastern Grey Kangaroo Macropodidae Macropus rufogriseus Red Necked Wallaby Macropodidae Wallabia bicolor Swamp Wallaby Miniopteridae Miniopterus schreibersii oceanensis ~ Eastern Bent-wing Bat Muridae Hydromys chrysogaster Water Rat Muridae Mus musculus ^ House Mouse Muridae Rattus fuscipes Bush Rat Muridae Rattus lutreolus Swamp Rat Otariidae Arctocephalus pusillus doriferus ~ Australian Fur-seal Otariidae Arctocephalus forsteri ~ New Zealand Fur Seal Peramelidae Isoodon obesulus Southern Brown Bandicoot Peramelidae Perameles nasuta Long-nosed Bandicoot Petauridae Petaurus australis Yellow Bellied Glider Petauridae Petaurus breviceps Sugar Glider Phalangeridae Trichosurus cunninghami Mountain Brushtail Possum Phalangeridae Trichosurus vulpecula Common Brushtail Possum Phascolarctidae Phascolarctos cinereus Koala Potoroidae Potorous sp. # ~ Long-nosed or Long-footed Potoroo Pseudocheiridae Petauroides volans Greater Glider Pseudocheiridae Pseudocheirus peregrinus -

The Sex Pheromone of the Sand Sagebrush
The Sex Pheromone of the Sand Sagebrush Carpenterworm, Holcocerus artemisiae (Lepidoptera, Cossidae) Jintong Zhanga,*, Xiaoyuan Jinga, Youqing Luob, Zhanwen Lic, Shixiang Zongb, and Meihong Yanga a Shanxi Agricultural University, Shanxi, 030801, China. Fax: (03 54-)6 28 69 90. E-mail: [email protected] b Beijing Forestry University, Beijing, 00083, China c Lingwu Station of Forest Diseases and Pests Control, Ningxia, 750001, China * Author for correspondence and reprint requests Z. Naturforsch. 64 c, 590 – 596 (2009); received November 10, 2008/January 12, 2009 (Z)-5-dodecen-1-ol (Z5 – 12:OH), (Z)-5-dodecenyl acetate (Z5 – 12:Ac), and (Z)-5-tetrade- cenyl acetate (Z5 – 14:Ac) were found in the extracts of the female sex pheromone gland of the carpenterworm moth Holcocerus artemisiae Chou et Hua, a pest of Artemisia fi lifolia. The average amounts of Z5 – 12:OH, Z5 – 12:Ac, and Z5 – 14:Ac in a single sex pheromone gland of a calling moth were (7.14 ± 0.73) ng, (54.20 ± 0.34) ng, and (38.70 ± 0.46) ng, re- spectively. Electroantennography (EAG) of these compounds and their analogues demon- strated that Z5 – 12:Ac excitated the largest male EAG response, followed by Z5 – 14:Ac. Traps baited with rubber septa impregnated with Z5 – 12:Ac (500 μg/septum) and Z5 – 14:Ac (300 μg/septum) were more effective than traps with other baits or virgin females. Addition of Z5 – 12:OH to rubber septa did not enhance the trap catches, but (E,Z)-3,5-dodecadienyl acetate (E3,Z5 – 12:Ac) enhanced the trap catch.